| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,931,609 – 7,931,762 |
| Length | 153 |
| Max. P | 0.999821 |

| Location | 7,931,609 – 7,931,723 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.35 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -24.78 |
| Energy contribution | -25.34 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.16 |
| SVM RNA-class probability | 0.999821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7931609 114 + 27905053 GUUCUUUUUUU------UCCUUUUUGUGGGGAAUUUAUUGCCUGAAGUUAUUAGCUUUGUAAUCUUUUAAUAAAGUUUUGCCCCAAUUGAUGGGGCUUGACAUCACUUGUACAGAGCAAC .........((------((((......))))))....................((((((((...(((....)))(((..((((((.....))))))..)))........))))))))... ( -28.20) >DroSec_CAF1 63190 112 + 1 GCUCUUUUUUU------UCC--UUUGUUGGGAAUUUAUUGCCUUAAGUUAUUAGCUUUGUAAUCUUUUAAUAAAGUUUUGUCCCAAUUGAUGGGGCUUUACAUCACUUGUACAGAGCAAC (((((......------..(--((((((((((....(((((...((((.....)))).))))).)))))))))))....((((((.....))))))..((((.....)))).)))))... ( -26.50) >DroSim_CAF1 65261 112 + 1 GCUCUUUUUUU------CCU--UUUGUUGGGAAUUUAUUGCCUUAAGUUAUUAGCUUUGUAAUCUUUUAAUAAAGUUUUGCCCCAAUUGAUGGGGCUUUACAUCACUUGUACAGAGCAAC (((((....((------(((--......)))))..........(((((....((((((((.........))))))))..((((((.....))))))........)))))...)))))... ( -27.80) >DroEre_CAF1 64108 115 + 1 GCUCUUUUUUUUUUG-----CUUUUGUUUGGAAUUUAUUGCCUGAAGUUAUUAGCUUUGUAAUCUUUUAAUAAAGUUUUGCCCCAAUUGAUGGGGCUUUACAUCACUUGUGCGGAGCAAA (((((.........(-----((((...((((((...(((((..(((((.....)))))))))).)))))).)))))...((((((.....))))))..((((.....)))).)))))... ( -26.90) >DroYak_CAF1 65513 120 + 1 GCUCUUUUUCUUUUCUUUCCUUUUUUUUUGGAAUUUAUUGCCUGAAGCUAUUUGCUUUGUAAUCUUUUAAUAAAGUUUUGCCCCAAUUGAUGGGGCUUUACAUCACUUGUGCAGAGCAAA ((((............((((.........)))).....(((..(((((.....)))))................((...((((((.....))))))...)).........)))))))... ( -28.50) >consensus GCUCUUUUUUU______UCC_UUUUGUUGGGAAUUUAUUGCCUGAAGUUAUUAGCUUUGUAAUCUUUUAAUAAAGUUUUGCCCCAAUUGAUGGGGCUUUACAUCACUUGUACAGAGCAAC (((((.................((((((((((....(((((..(((((.....)))))))))).))))))))))((...((((((.....))))))...))...........)))))... (-24.78 = -25.34 + 0.56)


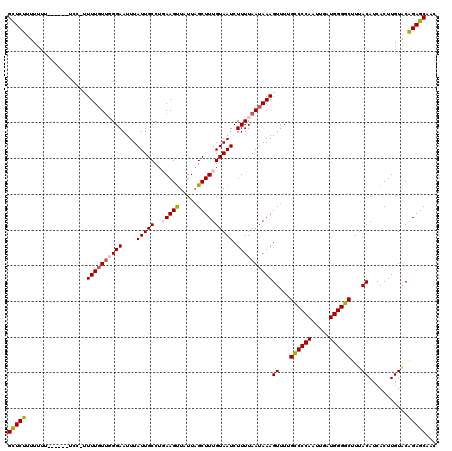
| Location | 7,931,609 – 7,931,723 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.35 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -19.62 |
| Energy contribution | -20.42 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7931609 114 - 27905053 GUUGCUCUGUACAAGUGAUGUCAAGCCCCAUCAAUUGGGGCAAAACUUUAUUAAAAGAUUACAAAGCUAAUAACUUCAGGCAAUAAAUUCCCCACAAAAAGGA------AAAAAAAGAAC (((((.(((....(((..(((...((((((.....))))))....((((....))))...)))..)))........))))))))...((((.........)))------).......... ( -24.40) >DroSec_CAF1 63190 112 - 1 GUUGCUCUGUACAAGUGAUGUAAAGCCCCAUCAAUUGGGACAAAACUUUAUUAAAAGAUUACAAAGCUAAUAACUUAAGGCAAUAAAUUCCCAACAAA--GGA------AAAAAAAGAGC ...(((((.....(((..(((((.(.((((.....)))).)....((((....)))).)))))..)))...................((((.......--)))------).....))))) ( -20.20) >DroSim_CAF1 65261 112 - 1 GUUGCUCUGUACAAGUGAUGUAAAGCCCCAUCAAUUGGGGCAAAACUUUAUUAAAAGAUUACAAAGCUAAUAACUUAAGGCAAUAAAUUCCCAACAAA--AGG------AAAAAAAGAGC ...(((((.....(((..(((((.((((((.....))))))....((((....)))).)))))..)))...................((((.......--.))------))....))))) ( -27.60) >DroEre_CAF1 64108 115 - 1 UUUGCUCCGCACAAGUGAUGUAAAGCCCCAUCAAUUGGGGCAAAACUUUAUUAAAAGAUUACAAAGCUAAUAACUUCAGGCAAUAAAUUCCAAACAAAAG-----CAAAAAAAAAAGAGC ((((((.(((....))).(((((.((((((.....))))))....((((....)))).)))))..(((..........))).................))-----))))........... ( -23.50) >DroYak_CAF1 65513 120 - 1 UUUGCUCUGCACAAGUGAUGUAAAGCCCCAUCAAUUGGGGCAAAACUUUAUUAAAAGAUUACAAAGCAAAUAGCUUCAGGCAAUAAAUUCCAAAAAAAAAGGAAAGAAAAGAAAAAGAGC ...(((((((....(((((.....((((((.....))))))....((((....))))))))).((((.....))))...))......((((.........))))...........))))) ( -29.30) >consensus GUUGCUCUGUACAAGUGAUGUAAAGCCCCAUCAAUUGGGGCAAAACUUUAUUAAAAGAUUACAAAGCUAAUAACUUCAGGCAAUAAAUUCCCAACAAAA_GGA______AAAAAAAGAGC ...(((((......(((((.....((((((.....))))))....((((....)))))))))...(((..........)))..................................))))) (-19.62 = -20.42 + 0.80)



| Location | 7,931,643 – 7,931,762 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.35 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -21.80 |
| Energy contribution | -22.08 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7931643 119 + 27905053 CCUGAAGUUAUUAGCUUUGUAAUCUUUUAAUAAAGUUUUGCCCCAAUUGAUGGGGCUUGACAUCACUUGUACAGAGCAACAAUGAUUU-UAGUUUUUGCCAUCAACUGAGUAAACAUUCU .(((((((((((.((((((((...(((....)))(((..((((((.....))))))..)))........))))))))...))))))))-)))..(((((((.....)).)))))...... ( -32.40) >DroSec_CAF1 63222 101 + 1 CCUUAAGUUAUUAGCUUUGUAAUCUUUUAAUAAAGUUUUGUCCCAAUUGAUGGGGCUUUACAUCACUUGUACAGAGCAACAAUGAUUU-UAGUUA---GCAACAA--------------- .((.((((((((.((((((((.........((((((((..((......))..)))))))).........))))))))...))))))))-.))...---.......--------------- ( -23.57) >DroSim_CAF1 65293 120 + 1 CCUUAAGUUAUUAGCUUUGUAAUCUUUUAAUAAAGUUUUGCCCCAAUUGAUGGGGCUUUACAUCACUUGUACAGAGCAACAAUGAUUUUUAGUUUUUGCCAUCAACUGAGUAAACAUUCU .((.((((((((.((((((((..((((....))))....((((((.....)))))).............))))))))...))))))))..))..(((((((.....)).)))))...... ( -25.90) >DroEre_CAF1 64143 119 + 1 CCUGAAGUUAUUAGCUUUGUAAUCUUUUAAUAAAGUUUUGCCCCAAUUGAUGGGGCUUUACAUCACUUGUGCGGAGCAAAAUUGAUUU-UGGUUUUGGCCAUCAGUUGUGUAAAUAUUUU .((((........((((((((..((((....))))....((((((.....)))))).............))))))))...........-((((....))))))))............... ( -27.80) >DroYak_CAF1 65553 119 + 1 CCUGAAGCUAUUUGCUUUGUAAUCUUUUAAUAAAGUUUUGCCCCAAUUGAUGGGGCUUUACAUCACUUGUGCAGAGCAAAAUUGAUUU-UAGUUUUGCCCAUCAAUUGUGUAAAUAUUUU ...(((((.....))))).............((((((((((..((((((((((((((((.(((.....))).)))))(((((((....-))))))).))))))))))).))))).))))) ( -31.20) >consensus CCUGAAGUUAUUAGCUUUGUAAUCUUUUAAUAAAGUUUUGCCCCAAUUGAUGGGGCUUUACAUCACUUGUACAGAGCAACAAUGAUUU_UAGUUUUGGCCAUCAACUGAGUAAACAUUCU .(((((((((((.((((((((..((((....))))....((((((.....)))))).............))))))))...)))))))).)))............................ (-21.80 = -22.08 + 0.28)


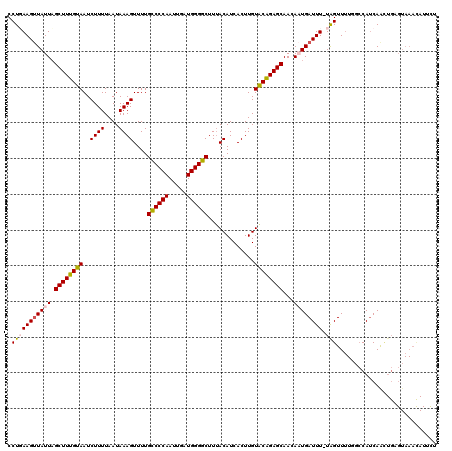
| Location | 7,931,643 – 7,931,762 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.35 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -16.66 |
| Energy contribution | -18.02 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7931643 119 - 27905053 AGAAUGUUUACUCAGUUGAUGGCAAAAACUA-AAAUCAUUGUUGCUCUGUACAAGUGAUGUCAAGCCCCAUCAAUUGGGGCAAAACUUUAUUAAAAGAUUACAAAGCUAAUAACUUCAGG ..........((.(((((.((((........-......((((........))))(((((.....((((((.....))))))....((((....)))))))))...)))).)))))..)). ( -26.50) >DroSec_CAF1 63222 101 - 1 ---------------UUGUUGC---UAACUA-AAAUCAUUGUUGCUCUGUACAAGUGAUGUAAAGCCCCAUCAAUUGGGACAAAACUUUAUUAAAAGAUUACAAAGCUAAUAACUUAAGG ---------------(((((((---(.....-..((((((((........)).))))))((((.(.((((.....)))).)....((((....)))).))))..))).)))))....... ( -15.90) >DroSim_CAF1 65293 120 - 1 AGAAUGUUUACUCAGUUGAUGGCAAAAACUAAAAAUCAUUGUUGCUCUGUACAAGUGAUGUAAAGCCCCAUCAAUUGGGGCAAAACUUUAUUAAAAGAUUACAAAGCUAAUAACUUAAGG .............(((((.((((...........((((((((........)).))))))((((.((((((.....))))))....((((....)))).))))...)))).)))))..... ( -27.30) >DroEre_CAF1 64143 119 - 1 AAAAUAUUUACACAACUGAUGGCCAAAACCA-AAAUCAAUUUUGCUCCGCACAAGUGAUGUAAAGCCCCAUCAAUUGGGGCAAAACUUUAUUAAAAGAUUACAAAGCUAAUAACUUCAGG ...............(((((((......)))-.((((...(((((..(((....)))..)))))((((((.....))))))...............))))...............)))). ( -26.10) >DroYak_CAF1 65553 119 - 1 AAAAUAUUUACACAAUUGAUGGGCAAAACUA-AAAUCAAUUUUGCUCUGCACAAGUGAUGUAAAGCCCCAUCAAUUGGGGCAAAACUUUAUUAAAAGAUUACAAAGCAAAUAGCUUCAGG ......((((((((.(((..((((((((...-.......))))))))....))).)).))))))((((((.....))))))....((((....))))......((((.....)))).... ( -28.30) >consensus AAAAUAUUUACACAAUUGAUGGCAAAAACUA_AAAUCAUUGUUGCUCUGUACAAGUGAUGUAAAGCCCCAUCAAUUGGGGCAAAACUUUAUUAAAAGAUUACAAAGCUAAUAACUUCAGG ...................((((...........((((((..(((...)))..))))))((((.((((((.....))))))....((((....)))).))))...))))........... (-16.66 = -18.02 + 1.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:10 2006