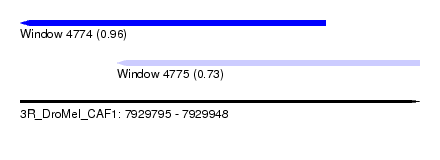
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,929,795 – 7,929,948 |
| Length | 153 |
| Max. P | 0.960246 |

| Location | 7,929,795 – 7,929,912 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -31.76 |
| Consensus MFE | -24.04 |
| Energy contribution | -23.56 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7929795 117 - 27905053 UUCUGGAAUUGGUUUAGGCCACUACAAAAUGAACAACCAUUUUGGACAUUAGGCCAAAGCUCAUCAGCAUCCCGAAUGUCAUCAGAAAUAGCCAUAGUUA--GAAAAUGGUCU-UACUAA ((((((...((((....))))...(((((((......)))))))((((((.((.....(((....)))...)).)))))).)))))).......((((.(--((......)))-.)))). ( -27.00) >DroSec_CAF1 61381 119 - 1 UUCUGGAAUUGGUUUAGGCCACUACAAAAUGAACAACCAUUUUGGACAUUAGACCAAAGCUCAUCAGCAUCCCGAAUGUCAUCAGAAAUGGCGAUAGCCAAUGACAAUGGGUU-UACUAA (((.(((.(((((((((.......(((((((......)))))))....))))))))).(((....))).))).)))((((((......((((....)))))))))).(((...-..))). ( -32.00) >DroSim_CAF1 63212 119 - 1 UUCUGGAAUUGGUUUAGGCCACUACAAAAUGAACAACCAUUUUGGACAUUUGGCCAAAGCUCAUCAGCAUCCCGAAUGUCAUCAGAAAUGGCGAUAGCCGAUGACAAUGGGUU-UACUAA ..((((....(((((.(((((...(((((((......)))))))......))))).)))))..))))...((((..((((((((....)(((....)))))))))).))))..-...... ( -37.00) >DroEre_CAF1 62329 108 - 1 UUCUGCAAUUGGUUUAGGCCACUACAAAAUGAACAACCAUUUUGGACAUUAGGCCAAAGCUCAUCGGCAUCCCCAAUGUCAUCAGAAAUGGCGAUAGUCG--GA----------UACUAA ....((...((((((.((((....(((((((......))))))).......)))).)))).))...))((((....((((..((....))..))))...)--))----------)..... ( -29.80) >DroYak_CAF1 63685 118 - 1 UUCUGGAAUUGGUUUAGGCCACUACAAAAUGAACAACCAUUUCGGACAUUAGGCCAAAGCUCAUAAGCAUCCCCGAUGUCAUCGGUAGUGGCGAUAGUCA--GACAAUGGGCUAUACUAA .............((((((((((((.(((((......)))))..(((((..((.....(((....)))...))..)))))....)))))))).((((((.--.......)))))).)))) ( -33.00) >consensus UUCUGGAAUUGGUUUAGGCCACUACAAAAUGAACAACCAUUUUGGACAUUAGGCCAAAGCUCAUCAGCAUCCCGAAUGUCAUCAGAAAUGGCGAUAGUCA__GACAAUGGGCU_UACUAA ((((((...((((....))))...(((((((......)))))))((((((.((.....(((....)))...)).)))))).)))))).((((....)))).................... (-24.04 = -23.56 + -0.48)



| Location | 7,929,832 – 7,929,948 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.63 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -21.18 |
| Energy contribution | -22.54 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.727006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7929832 116 - 27905053 AACAAGCAUGCUGGCCAUGCCC----AGCAGUUAUGUAAAUUCUGGAAUUGGUUUAGGCCACUACAAAAUGAACAACCAUUUUGGACAUUAGGCCAAAGCUCAUCAGCAUCCCGAAUGUC .((((((.((((((......))----))))))).)))..((((.(((.(((((((((.......(((((((......)))))))....))))))))).(((....))).))).))))... ( -33.60) >DroSec_CAF1 61420 116 - 1 AACAAGCAUGCUCGCCAUGCCC----GGCAGUUAUGUAAAUUCUGGAAUUGGUUUAGGCCACUACAAAAUGAACAACCAUUUUGGACAUUAGACCAAAGCUCAUCAGCAUCCCGAAUGUC .....(((((((.(((......----))))).)))))..((((.(((.(((((((((.......(((((((......)))))))....))))))))).(((....))).))).))))... ( -31.60) >DroSim_CAF1 63251 116 - 1 AACAAGCAUGCUCGCCAUGCCC----AGCAGUUAUGUAAAUUCUGGAAUUGGUUUAGGCCACUACAAAAUGAACAACCAUUUUGGACAUUUGGCCAAAGCUCAUCAGCAUCCCGAAUGUC .....((((..(((..((((((----((.((((.....))))))))....(((((.(((((...(((((((......)))))))......))))).))))).....))))..))))))). ( -30.50) >DroEre_CAF1 62357 120 - 1 AGCAAGCAUGCUCACCAUGCCCAGCCAGCAGUUAUGCAAAUUCUGCAAUUGGUUUAGGCCACUACAAAAUGAACAACCAUUUUGGACAUUAGGCCAAAGCUCAUCGGCAUCCCCAAUGUC .(((.(((((.....)))))...(((.((((...........))))....(((((.((((....(((((((......))))))).......)))).)))))....)))........))). ( -31.60) >DroYak_CAF1 63723 103 - 1 -------------ACCAUGCCC----AGCAGUUAUGUAAAUUCUGGAAUUGGUUUAGGCCACUACAAAAUGAACAACCAUUUCGGACAUUAGGCCAAAGCUCAUAAGCAUCCCCGAUGUC -------------((((...((----((.((((.....))))))))...))))...((((....(.(((((......))))).).......))))...........(((((...))))). ( -20.30) >consensus AACAAGCAUGCUCGCCAUGCCC____AGCAGUUAUGUAAAUUCUGGAAUUGGUUUAGGCCACUACAAAAUGAACAACCAUUUUGGACAUUAGGCCAAAGCUCAUCAGCAUCCCGAAUGUC .....(((((.....))))).......(((....)))..((((.(((.(((((((((.......(((((((......)))))))....))))))))).(((....))).))).))))... (-21.18 = -22.54 + 1.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:59 2006