
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,929,014 – 7,929,167 |
| Length | 153 |
| Max. P | 0.998271 |

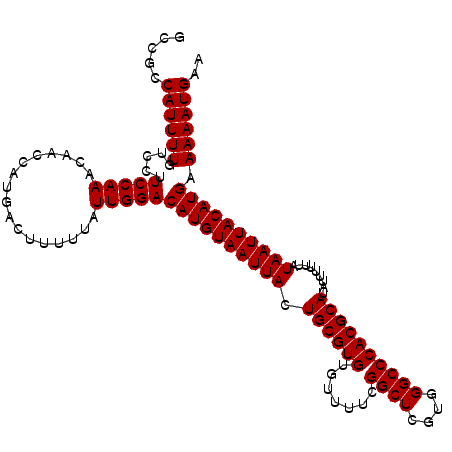
| Location | 7,929,014 – 7,929,127 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 98.05 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -25.54 |
| Energy contribution | -25.54 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7929014 113 + 27905053 GCCGCCAUUUUGUCCUUCCAAACAACCAUGACAUUUUAUUGGACAUGUAAUUACUGCGUGUGUUUUCGGCUCGCGGGCCCACGCAUAUUUUUUAUAAUUACAUGAAAAAUGAA .....((((((.....(((((.................)))))((((((((((.((((((.......((((....)))))))))).........)))))))))).)))))).. ( -25.54) >DroSec_CAF1 60191 113 + 1 GCCGCCAUUUUGUCCUUCCAAACAACCAUGAAUUUUUAUUGGACAUGUAAUUACUGCGUGUGUUUUCGGCUCGUGGGCCCACGCAUAUUUUUUAUAAUUACAUGAAAAAUGAA .....((((((.....(((((.................)))))((((((((((.((((((.......((((....)))))))))).........)))))))))).)))))).. ( -25.54) >DroSim_CAF1 62002 113 + 1 GCCGCCAUUUUGUCCUUCCAAACAACCAUGACUUUUUAUUGGACAUGUAAUUACUGCGUGUGUUUUCGGCUCGCGGGCCCACGCAUAUUUUUUAUAAUUACAUGAAAAAUGAA .....((((((.....(((((.................)))))((((((((((.((((((.......((((....)))))))))).........)))))))))).)))))).. ( -25.54) >DroEre_CAF1 61077 112 + 1 GCCGCCAUUUUGUCCUUCCAAACAACCAUGACUUUUUAUUGGACAUGUAAUUACUGCGUGCGUUUUCGGCUCGUGGGCCCACGCAUAUUU-UUAUAAUUACAUGAAAAAUGAA .....((((((.....(((((.................)))))((((((((((....((((((....((((....)))).))))))....-...)))))))))).)))))).. ( -26.83) >DroYak_CAF1 62387 113 + 1 GCCGCCAUUUUGUCCUUCCAAACAACCAUGACUUUUUAUUGGACAUGUAAUUACUGCGUGUGUUUUCGGCUCGUGGGCCCACGCAUAUUUUUUAUAAUUACAUGAAAAAUGAA .....((((((.....(((((.................)))))((((((((((.((((((.......((((....)))))))))).........)))))))))).)))))).. ( -25.54) >consensus GCCGCCAUUUUGUCCUUCCAAACAACCAUGACUUUUUAUUGGACAUGUAAUUACUGCGUGUGUUUUCGGCUCGUGGGCCCACGCAUAUUUUUUAUAAUUACAUGAAAAAUGAA .....((((((.....(((((.................)))))((((((((((.((((((.......((((....)))))))))).........)))))))))).)))))).. (-25.54 = -25.54 + -0.00)


| Location | 7,929,014 – 7,929,127 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 98.05 |
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -28.42 |
| Energy contribution | -28.62 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7929014 113 - 27905053 UUCAUUUUUCAUGUAAUUAUAAAAAAUAUGCGUGGGCCCGCGAGCCGAAAACACACGCAGUAAUUACAUGUCCAAUAAAAUGUCAUGGUUGUUUGGAAGGACAAAAUGGCGGC .........(((((((((((........(((((((((......)))(....).)))))))))))))))))..........((((((..((((((....)))))).)))))).. ( -29.80) >DroSec_CAF1 60191 113 - 1 UUCAUUUUUCAUGUAAUUAUAAAAAAUAUGCGUGGGCCCACGAGCCGAAAACACACGCAGUAAUUACAUGUCCAAUAAAAAUUCAUGGUUGUUUGGAAGGACAAAAUGGCGGC .(((((((.(((((((((((........(((((((((......)))(....).)))))))))))))))))(((((....(((.....)))..))))).....))))))).... ( -27.10) >DroSim_CAF1 62002 113 - 1 UUCAUUUUUCAUGUAAUUAUAAAAAAUAUGCGUGGGCCCGCGAGCCGAAAACACACGCAGUAAUUACAUGUCCAAUAAAAAGUCAUGGUUGUUUGGAAGGACAAAAUGGCGGC .........(((((((((((........(((((((((......)))(....).)))))))))))))))))...........(((((..((((((....)))))).)))))... ( -29.70) >DroEre_CAF1 61077 112 - 1 UUCAUUUUUCAUGUAAUUAUAA-AAAUAUGCGUGGGCCCACGAGCCGAAAACGCACGCAGUAAUUACAUGUCCAAUAAAAAGUCAUGGUUGUUUGGAAGGACAAAAUGGCGGC .........(((((((((((..-.....(((((((((......)))(....).)))))))))))))))))...........(((((..((((((....)))))).)))))... ( -29.51) >DroYak_CAF1 62387 113 - 1 UUCAUUUUUCAUGUAAUUAUAAAAAAUAUGCGUGGGCCCACGAGCCGAAAACACACGCAGUAAUUACAUGUCCAAUAAAAAGUCAUGGUUGUUUGGAAGGACAAAAUGGCGGC .........(((((((((((........(((((((((......)))(....).)))))))))))))))))...........(((((..((((((....)))))).)))))... ( -29.70) >consensus UUCAUUUUUCAUGUAAUUAUAAAAAAUAUGCGUGGGCCCACGAGCCGAAAACACACGCAGUAAUUACAUGUCCAAUAAAAAGUCAUGGUUGUUUGGAAGGACAAAAUGGCGGC .........(((((((((((........(((((((((......)))(....).)))))))))))))))))...........(((((..((((((....)))))).)))))... (-28.42 = -28.62 + 0.20)



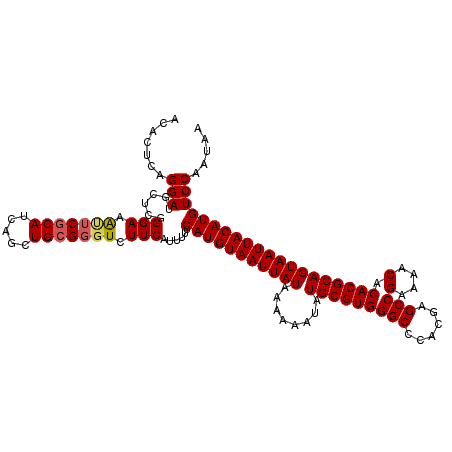
| Location | 7,929,049 – 7,929,167 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 96.19 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -30.30 |
| Energy contribution | -30.62 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7929049 118 - 27905053 ACACUCAGGAUGCUCGGGAAAUUCGCAUCAGCUGGGGGUCUUCAUUUUUCAUGUAAUUAUAAAAAAUAUGCGUGGGCCCGCGAGCCGAAAACACACGCAGUAAUUACAUGUCCAAUAA ...((((((((((...........)))))..))))).............(((((((((((........(((((((((......)))(....).)))))))))))))))))........ ( -30.90) >DroSec_CAF1 60226 118 - 1 ACAGUCAGGAUGCUCCGGAAAUUCGCAUCAGCUGCGGGUCUUCAUUUUUCAUGUAAUUAUAAAAAAUAUGCGUGGGCCCACGAGCCGAAAACACACGCAGUAAUUACAUGUCCAAUAA .......(((......(((.(((((((.....))))))).)))......(((((((((((........(((((((((......)))(....).))))))))))))))))))))..... ( -32.70) >DroSim_CAF1 62037 118 - 1 ACACUCAGGAUGCUCGGGAAAUUCGCAUCAGCUGCGGGUCUUCAUUUUUCAUGUAAUUAUAAAAAAUAUGCGUGGGCCCGCGAGCCGAAAACACACGCAGUAAUUACAUGUCCAAUAA .......(((......(((.(((((((.....))))))).)))......(((((((((((........(((((((((......)))(....).))))))))))))))))))))..... ( -32.40) >DroEre_CAF1 61112 116 - 1 ACACUCAGGAUGCUCGGGAAG-UCGCAUCAGCUGCGGGUCUUCAUUUUUCAUGUAAUUAUAA-AAAUAUGCGUGGGCCCACGAGCCGAAAACGCACGCAGUAAUUACAUGUCCAAUAA .......(((.......((((-(((((.....)))))..))))......(((((((((((..-.....(((((((((......)))(....).))))))))))))))))))))..... ( -31.31) >DroYak_CAF1 62422 118 - 1 UCACUCAGGAUGCUCGGGAAACGCGCAUCAGCUGCGGGUCUUCAUUUUUCAUGUAAUUAUAAAAAAUAUGCGUGGGCCCACGAGCCGAAAACACACGCAGUAAUUACAUGUCCAAUAA .......(((.....((((..(((((....)).)))..)))).......(((((((((((........(((((((((......)))(....).))))))))))))))))))))..... ( -33.30) >consensus ACACUCAGGAUGCUCGGGAAAUUCGCAUCAGCUGCGGGUCUUCAUUUUUCAUGUAAUUAUAAAAAAUAUGCGUGGGCCCACGAGCCGAAAACACACGCAGUAAUUACAUGUCCAAUAA .......(((......(((.(((((((.....))))))).)))......(((((((((((........(((((((((......)))(....).))))))))))))))))))))..... (-30.30 = -30.62 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:57 2006