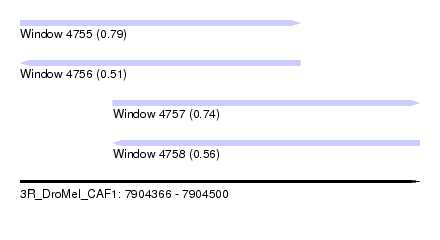
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,904,366 – 7,904,500 |
| Length | 134 |
| Max. P | 0.791502 |

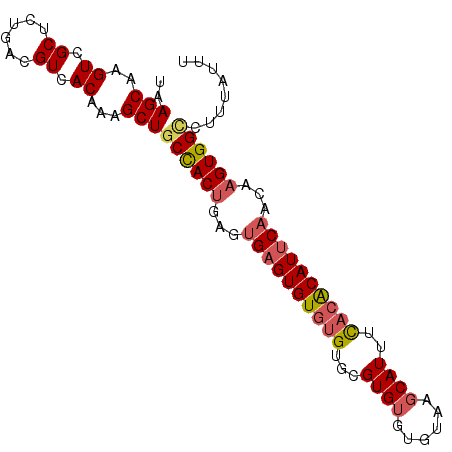
| Location | 7,904,366 – 7,904,460 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 88.83 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -23.11 |
| Energy contribution | -24.18 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7904366 94 + 27905053 UAAGCAAGUCGCUCUGGCGUCACAAAGCUUCUACUGGGUGAGUGUGUGUGCGUGUGUGUAAGCAUUUUACACAUUCAACAAGUGGCCUUUAUUU .......(.(((....))).)..((((...(((((...((((((((((...((((......))))..))))))))))...))))).)))).... ( -27.00) >DroSec_CAF1 35764 92 + 1 UAAGCAAGUCGCUCUGACGUCACAAUGCUGCCACUGAGUGAGUGUGUGUGCGUG--UGUAAGCAUUUCACACAUUCAACAAGUGGCCUUUAUUU ..((((.((.((......)).))..))))((((((...((((((((((...(((--(....))))..))))))))))...))))))........ ( -32.70) >DroSim_CAF1 37503 94 + 1 UAAGCAAGUCGCUCUGACGUCACAAAGCUGCCACUGAGUGAGUGUGUGUGCGUGUGUGUAAGCAUUUCACACAUUCAACAAGUGGCCUUUAUUU ..........(((.((......)).))).((((((...((((((((((...((((......))))..))))))))))...))))))........ ( -32.10) >DroYak_CAF1 36377 92 + 1 UAAGCAAGUCGCUCUGACGUCACUAAGCUGCCACAGAGAGUGUGUGCGUGCGUGUGAGUGAGCAUUCC--GCAUUCAACAGGUGGUCUUUAUUU ..........((((..((.((((...(((((((((.....)))).))).))..))))))))))...((--((.........))))......... ( -23.20) >consensus UAAGCAAGUCGCUCUGACGUCACAAAGCUGCCACUGAGUGAGUGUGUGUGCGUGUGUGUAAGCAUUUCACACAUUCAACAAGUGGCCUUUAUUU ..(((..((.((......)).))...)))((((((...((((((((((...((((......))))..))))))))))...))))))........ (-23.11 = -24.18 + 1.06)

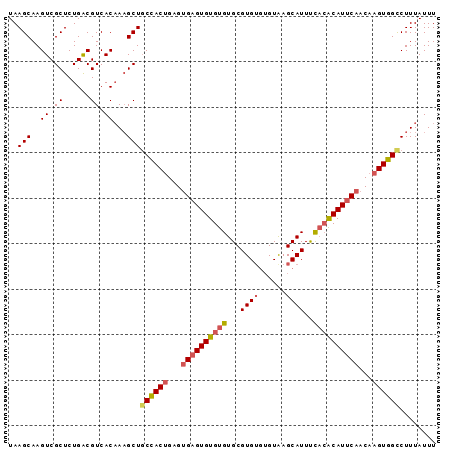
| Location | 7,904,366 – 7,904,460 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 88.83 |
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -16.99 |
| Energy contribution | -19.80 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7904366 94 - 27905053 AAAUAAAGGCCACUUGUUGAAUGUGUAAAAUGCUUACACACACGCACACACACUCACCCAGUAGAAGCUUUGUGACGCCAGAGCGACUUGCUUA ......((((.(((.(.(((.(((((....(((..........))).))))).))).).)))((..((((((......))))))..)).)))). ( -19.70) >DroSec_CAF1 35764 92 - 1 AAAUAAAGGCCACUUGUUGAAUGUGUGAAAUGCUUACA--CACGCACACACACUCACUCAGUGGCAGCAUUGUGACGUCAGAGCGACUUGCUUA ........((((((.(.(((.((((((...(((.....--...))))))))).))).).))))))......(..(.(((.....))))..)... ( -26.40) >DroSim_CAF1 37503 94 - 1 AAAUAAAGGCCACUUGUUGAAUGUGUGAAAUGCUUACACACACGCACACACACUCACUCAGUGGCAGCUUUGUGACGUCAGAGCGACUUGCUUA .....(((((((((.(.(((.((((((...(((..........))))))))).))).).)))))).((((((......))))))..)))..... ( -29.80) >DroYak_CAF1 36377 92 - 1 AAAUAAAGACCACCUGUUGAAUGC--GGAAUGCUCACUCACACGCACGCACACACUCUCUGUGGCAGCUUAGUGACGUCAGAGCGACUUGCUUA ......................((--((..(((((((((((..((..((.((((.....)))))).))...)))).))..)))))..))))... ( -19.90) >consensus AAAUAAAGGCCACUUGUUGAAUGUGUGAAAUGCUUACACACACGCACACACACUCACUCAGUGGCAGCUUUGUGACGUCAGAGCGACUUGCUUA .....(((((((((.(.(((.((((((...(((..........))))))))).))).).)))))).((((((......))))))..)))..... (-16.99 = -19.80 + 2.81)

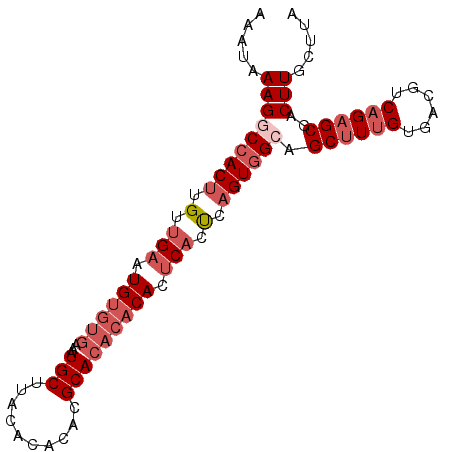

| Location | 7,904,397 – 7,904,500 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 90.29 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -23.83 |
| Energy contribution | -24.57 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7904397 103 + 27905053 UACUGGGUGAGUGUGUGUGCGUGUGUGUAAGCAUUUUACACAUUCAACAAGUGGCCUUUAUUUGGGCAACAAAGAAAAGGAAGCUGCAGCAAACUAAAGUUCG ..((((.((((((((((...((((......))))..)))))))))).).(((..(((((.((((.....))))..)))))..))).))).............. ( -27.90) >DroSec_CAF1 35795 101 + 1 CACUGAGUGAGUGUGUGUGCGUG--UGUAAGCAUUUCACACAUUCAACAAGUGGCCUUUAUUUGGGCAACAAAGAAAAGGAAGCUGCAGCAAAAUAAAGUUCU ((((...((((((((((...(((--(....))))..))))))))))...)))).(((((.((((.....))))..)))))..((....))............. ( -30.70) >DroSim_CAF1 37534 103 + 1 CACUGAGUGAGUGUGUGUGCGUGUGUGUAAGCAUUUCACACAUUCAACAAGUGGCCUUUAUUUGGGCAACAAAGAAAAGGAAGCUGCAGCAAAAUAAAGUUCU ((((...((((((((((...((((......))))..))))))))))...)))).(((((.((((.....))))..)))))..((....))............. ( -30.90) >DroYak_CAF1 36408 101 + 1 CACAGAGAGUGUGUGCGUGCGUGUGAGUGAGCAUUCC--GCAUUCAACAGGUGGUCUUUAUUUGGGCAACAAAGAAAAGGAAGCUGCAGCGAAAUAAAGUUCU (((...(((((((.(.((((..........)))).))--)))))).....)))..((((((((.(....)............((....)).)))))))).... ( -22.00) >consensus CACUGAGUGAGUGUGUGUGCGUGUGUGUAAGCAUUUCACACAUUCAACAAGUGGCCUUUAUUUGGGCAACAAAGAAAAGGAAGCUGCAGCAAAAUAAAGUUCU ((((...((((((((((...((((......))))..))))))))))...)))).(((((.((((.....))))..)))))..((....))............. (-23.83 = -24.57 + 0.75)


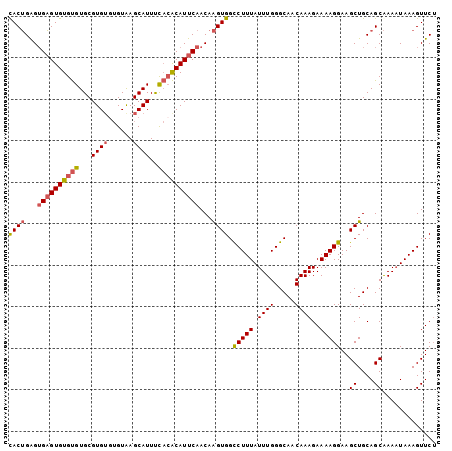
| Location | 7,904,397 – 7,904,500 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 90.29 |
| Mean single sequence MFE | -22.21 |
| Consensus MFE | -16.11 |
| Energy contribution | -18.18 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7904397 103 - 27905053 CGAACUUUAGUUUGCUGCAGCUUCCUUUUCUUUGUUGCCCAAAUAAAGGCCACUUGUUGAAUGUGUAAAAUGCUUACACACACGCACACACACUCACCCAGUA .(..(((((.((((..(((((............))))).)))))))))..)(((.(.(((.(((((....(((..........))).))))).))).).))). ( -20.60) >DroSec_CAF1 35795 101 - 1 AGAACUUUAUUUUGCUGCAGCUUCCUUUUCUUUGUUGCCCAAAUAAAGGCCACUUGUUGAAUGUGUGAAAUGCUUACA--CACGCACACACACUCACUCAGUG ....((((((((....(((((............)))))..))))))))..((((.(.(((.((((((...(((.....--...))))))))).))).).)))) ( -24.50) >DroSim_CAF1 37534 103 - 1 AGAACUUUAUUUUGCUGCAGCUUCCUUUUCUUUGUUGCCCAAAUAAAGGCCACUUGUUGAAUGUGUGAAAUGCUUACACACACGCACACACACUCACUCAGUG ....((((((((....(((((............)))))..))))))))..((((.(.(((.((((((...(((..........))))))))).))).).)))) ( -23.60) >DroYak_CAF1 36408 101 - 1 AGAACUUUAUUUCGCUGCAGCUUCCUUUUCUUUGUUGCCCAAAUAAAGACCACCUGUUGAAUGC--GGAAUGCUCACUCACACGCACGCACACACUCUCUGUG .......((((((((..((((.......((((((((.....))))))))......))))...))--))))))..................((((.....)))) ( -20.12) >consensus AGAACUUUAUUUUGCUGCAGCUUCCUUUUCUUUGUUGCCCAAAUAAAGGCCACUUGUUGAAUGUGUGAAAUGCUUACACACACGCACACACACUCACUCAGUG ....((((((((....(((((............)))))..))))))))..((((.(.(((.((((((...(((..........))))))))).))).).)))) (-16.11 = -18.18 + 2.06)


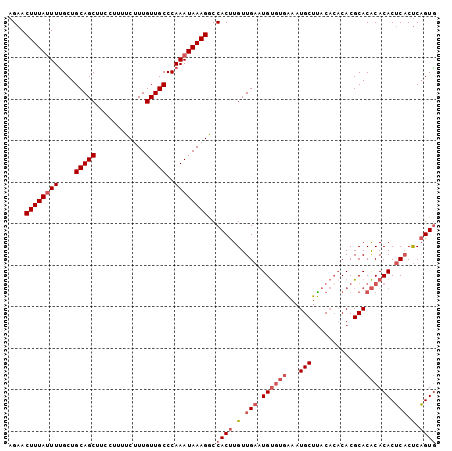
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:42 2006