| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
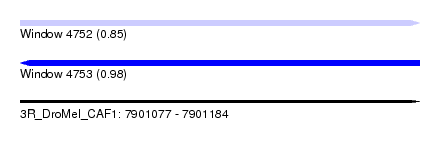
| Location | 7,901,077 – 7,901,184 |
| Length | 107 |
| Max. P | 0.984741 |

| Location | 7,901,077 – 7,901,184 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.69 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -17.64 |
| Energy contribution | -17.78 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7901077 107 + 27905053 UAUUGGUGGUCGGAUAGUACAUCUCCUCAUCCCAGUUGAUGAGGAACGACUGUACAAAGCGUAUAACCUGUACGGA---AU-AUUGGUGUUUGUUUCUCAGAAUGGA--AUUU (((..(((.((.....(((((((((((((((......))))))))..)).)))))....(((((.....)))))))---.)-))..)))((..(((....)))..))--.... ( -27.70) >DroGri_CAF1 20377 104 + 1 UUUGGGUGCGUGCAUAGUACAUCUCCUCAUCCCAGUUGAUGAGGAACGAUUGAACAAAUCGUAUAACCUGCAGCAU---AUAU---AUACAUAU---UGAGAUGUAUACACAG .....((((.((((.........((((((((......))))))))((((((.....))))))......))))))))---...(---((((((..---....)))))))..... ( -30.50) >DroSec_CAF1 32501 104 + 1 UAUUGGUGGUCGGAUAGUACAUCUCCUCAUCCCAGUUGAUGAGGAACGACUGUACGAAGCGUAUAACCUGCACGGU---AU-U---AUAUUUGUUUCUCAGAAAGGC--AUUU ........(((.....(((((((((((((((......))))))))..)).)))))(((((((((((((.....)))---..-)---))))..))))).......)))--.... ( -27.50) >DroSim_CAF1 33459 104 + 1 UAUUGGUGGUCGGAUAGUACAUCUCCUCAUCCCAGUUGAUGAGGAACGACUGUACGAAGCGUAUAACCUGCACGGU---AU-U---AUAUUUGUUUCUCAGAAAGGC--AUUU ........(((.....(((((((((((((((......))))))))..)).)))))(((((((((((((.....)))---..-)---))))..))))).......)))--.... ( -27.50) >DroWil_CAF1 8917 99 + 1 UAUUGGUGCGUGCAUAAUACAUUUCCUCAUCCCAAUUGAUGAGGAACGACUGUACAAAGCGUAUUACCUGCCAUGAAAGAU-G---AUAUAUAUAGU----AAUG------GU ...((((((((......((((.(((((((((......)))))))))....))))....))))))))...(((((.....((-(---.....)))...----.)))------)) ( -23.60) >DroAna_CAF1 29194 95 + 1 UAUUGGUGGUCGCAUAGUACAUCUCCUCAUCCCAGUUAAUGAGGAACGAUUGUACAAAGCGGAUGACCUGGGGAA----AU-C---AUUAGCA--UC------GGGU--AUUG ..((((((..(((...((((((((((((((........)))))))..)).)))))...)))(((..((...))..----))-)---.....))--))------))..--.... ( -23.50) >consensus UAUUGGUGGUCGCAUAGUACAUCUCCUCAUCCCAGUUGAUGAGGAACGACUGUACAAAGCGUAUAACCUGCACGGU___AU_U___AUAUUUAUUUCUCAGAAAGGC__AUUU ....(((...((....(((((((((((((((......))))))))..)).)))))....))....)))............................................. (-17.64 = -17.78 + 0.14)

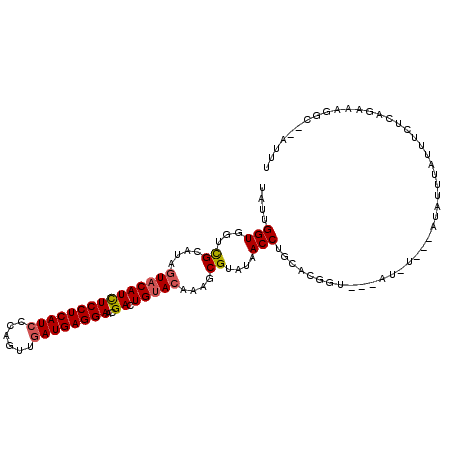
| Location | 7,901,077 – 7,901,184 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.69 |
| Mean single sequence MFE | -24.59 |
| Consensus MFE | -19.81 |
| Energy contribution | -19.56 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7901077 107 - 27905053 AAAU--UCCAUUCUGAGAAACAAACACCAAU-AU---UCCGUACAGGUUAUACGCUUUGUACAGUCGUUCCUCAUCAACUGGGAUGAGGAGAUGUACUAUCCGACCACCAAUA ....--.........................-..---........(((....((....(((((....(((((((((......))))))))).)))))....))...))).... ( -22.50) >DroGri_CAF1 20377 104 - 1 CUGUGUAUACAUCUCA---AUAUGUAU---AUAU---AUGCUGCAGGUUAUACGAUUUGUUCAAUCGUUCCUCAUCAACUGGGAUGAGGAGAUGUACUAUGCACGCACCCAAA .(((((((((((....---..))))))---))))---)(((((((...(((((((((.....)))))(((((((((......))))))))).))))...)))).)))...... ( -32.80) >DroSec_CAF1 32501 104 - 1 AAAU--GCCUUUCUGAGAAACAAAUAU---A-AU---ACCGUGCAGGUUAUACGCUUCGUACAGUCGUUCCUCAUCAACUGGGAUGAGGAGAUGUACUAUCCGACCACCAAUA ....--.....((.(((((.(...(((---(-..---(((.....))))))).).)))(((((....(((((((((......))))))))).)))))..)).))......... ( -22.50) >DroSim_CAF1 33459 104 - 1 AAAU--GCCUUUCUGAGAAACAAAUAU---A-AU---ACCGUGCAGGUUAUACGCUUCGUACAGUCGUUCCUCAUCAACUGGGAUGAGGAGAUGUACUAUCCGACCACCAAUA ....--.....((.(((((.(...(((---(-..---(((.....))))))).).)))(((((....(((((((((......))))))))).)))))..)).))......... ( -22.50) >DroWil_CAF1 8917 99 - 1 AC------CAUU----ACUAUAUAUAU---C-AUCUUUCAUGGCAGGUAAUACGCUUUGUACAGUCGUUCCUCAUCAAUUGGGAUGAGGAAAUGUAUUAUGCACGCACCAAUA .(------(((.----...........---.-.......))))..(((.....((...(((((....(((((((((......))))))))).)))))...))....))).... ( -23.35) >DroAna_CAF1 29194 95 - 1 CAAU--ACCC------GA--UGCUAAU---G-AU----UUCCCCAGGUCAUCCGCUUUGUACAAUCGUUCCUCAUUAACUGGGAUGAGGAGAUGUACUAUGCGACCACCAAUA ....--....------..--.....((---(-((----(......)))))).(((...(((((....(((((((((......))))))))).)))))...))).......... ( -23.90) >consensus AAAU__ACCAUUCUGAGAAACAAAUAU___A_AU___ACCGUGCAGGUUAUACGCUUUGUACAGUCGUUCCUCAUCAACUGGGAUGAGGAGAUGUACUAUCCGACCACCAAUA .............................................(((..........(((((....(((((((((......))))))))).))))).........))).... (-19.81 = -19.56 + -0.25)
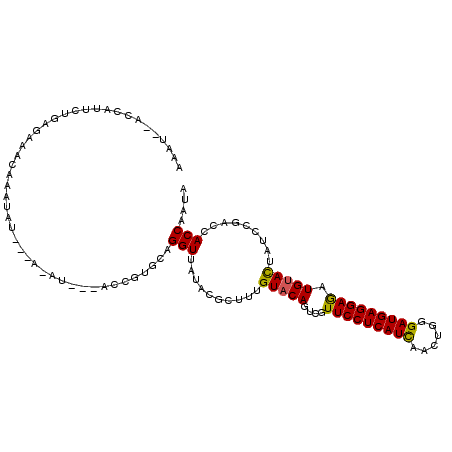
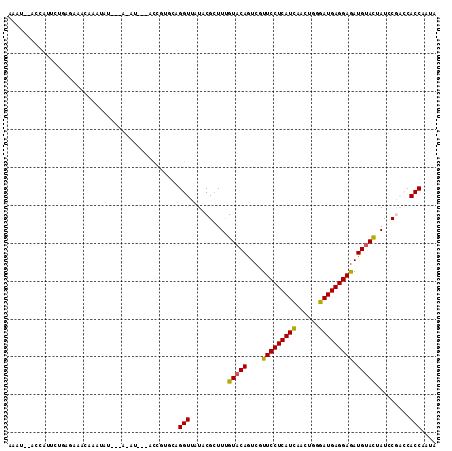
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:38 2006