
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,890,557 – 7,890,660 |
| Length | 103 |
| Max. P | 0.710157 |

| Location | 7,890,557 – 7,890,660 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -23.79 |
| Consensus MFE | -14.98 |
| Energy contribution | -15.12 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7890557 103 + 27905053 ACAAGUUGCAAGCUG----AAAACAUAG------GA-GCGAAAAGGUUGGCAGCAAUUUUGUUGCC-----UUAAUGCAUAAUAGAAAGUGUCGUAUCCAUGACAGCCA-AACUUUAAAA ..(((((....((((----....(((.(------((-((((.......(((((((....)))))))-----......(((........))))))).)))))).))))..-)))))..... ( -24.00) >DroPse_CAF1 21428 115 + 1 ACAAGUUGUAAGCAC----AACAAAAAAUGGCCAAA-GUAAAAAGGUUGAAAACAAUUUUGUUGCCGUUCCCUAAUGCAUAAUAGAAAGUGUCGUAUCCAUGGCAGCGAAAACUUUAAAA ....(((((....))----))).......((((...-.......))))........((((((((((((........((((........)))).......))))))))))))......... ( -21.56) >DroEre_CAF1 22394 103 + 1 ACAAGUUGCAAGCUG----AAAACAUAG------GA-GCGAAAAGGUUGGCAACAAUUUGGUUGCC-----UUAAUGCAUAAUAGAAAGUGUCGUAUCCAUGACAGCCA-AACUUUAAAA ..(((((....((((----......(((------(.-((((..((((((....))))))..)))))-----)))((((((........))).)))........))))..-)))))..... ( -23.80) >DroYak_CAF1 22536 102 + 1 ACAAGUUGCAAGCUG----GAAACAUAG------UA-GCGAA-AGGUUGGCAGCAAUUUGGUUGCC-----UUAAUGCAUAAUAGAAAGUGUCGUAUCCAUGACAGCCA-AACUUUAAAA ....(((((....((----....))..)------))-)).((-((.(((((.(((.(..((...))-----..).)))...........((((((....))))))))))-).)))).... ( -24.80) >DroAna_CAF1 19065 107 + 1 GCAAGUUGCAAGCAAAGGAAAAAAGUAU------GGGGCG-AAAGGUUGGAAACAAUUUUGUUGCC-----UUAAUGCAUAAUAGAAAGUGUCGCAUCCAUGGCAGCAA-AAGUUUAAAA ....(((((.......(((........(------((((((-((((((((....))))))).)))))-----))).((((((........))).))))))...)))))..-.......... ( -27.00) >DroPer_CAF1 21097 115 + 1 ACAAGUUGUAAGCAC----AACAAAAAAUGGCCAAA-GUAAAAAGGUUGAAAACAAUUUUGUUGCCGUUCCCUAAUGCAUAAUAGAAAGUGUCGUAUCCAUGGCAGCGAAAACUUUAAAA ....(((((....))----))).......((((...-.......))))........((((((((((((........((((........)))).......))))))))))))......... ( -21.56) >consensus ACAAGUUGCAAGCAG____AAAAAAUAG______GA_GCGAAAAGGUUGGAAACAAUUUUGUUGCC_____UUAAUGCAUAAUAGAAAGUGUCGUAUCCAUGACAGCCA_AACUUUAAAA ..(((((....((........................((((.(((((((....))))))).))))........................((((((....))))))))...)))))..... (-14.98 = -15.12 + 0.14)

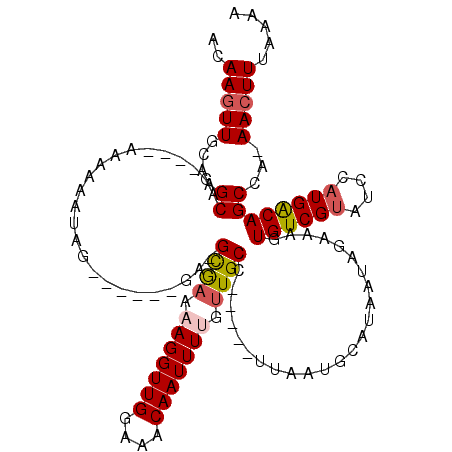
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:33 2006