
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,889,338 – 7,889,445 |
| Length | 107 |
| Max. P | 0.633559 |

| Location | 7,889,338 – 7,889,445 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 83.30 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -16.25 |
| Energy contribution | -17.26 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
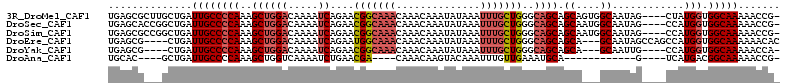
>3R_DroMel_CAF1 7889338 107 - 27905053 UGAGCGCUUGCUGAUUGCCCCAAAGCUGGACAAAAUCAGAACGGCAAACAAACAAAUAUAAAUUUGCUGGGCAGCAGCAGUGGCAAUAG----CUAUGGUGGCAAAAACCG- ((..(((((((((.((((((....((((..(.......)..)))).......(((((....)))))..)))))))))))(((((....)----)))))))).)).......- ( -30.60) >DroSec_CAF1 20798 107 - 1 UGAGCACCGGCUGAUUGCCCCAAAGCUGGACAAAAUCAGAACGGCAAACAAACAAAUAUAAAUUUGCUGGGCAGCAGCAAUGGCAAUAG----CCAUGGUGGCAAAAACCG- ((..((((.((((.((((((....((((..(.......)..)))).......(((((....)))))..)))))))))).(((((....)----)))))))).)).......- ( -35.20) >DroSim_CAF1 21226 107 - 1 UGAGCGCCGGCUGAUUGCCCCAAAGCUGGACAAAAUCAGAACGGCAAACAAACAAAUAUAAAUUUGCUGGGCAGCAGCAAUGGCAAUAG----CCAUGGUGGCAAAAACCG- .....((((((((.((((((....((((..(.......)..)))).......(((((....)))))..)))))))))).(((((....)----))))..))))........- ( -34.90) >DroEre_CAF1 21121 105 - 1 UGAGCG----CUGAUUGCCCCAAAGCUGGACAAAAUCAGAAUGGCAAACAAACAAAUAUAAAUUUGCUGGGCAGCAGCA---GCAAUAGCCAGCCAUGGUGGCAAAAAACAC ...(((----(((.((((((.....((((......))))...((((((..............)))))))))))))))).---))....((((.(....)))))......... ( -29.94) >DroYak_CAF1 21260 100 - 1 UGAGCG----CUGAUUGCCCCAAAGCUGGACAAAAUCAGAACGGCAAACAAACAAAUAUAAAUUUGCUGGGCAGCAGCA---GCAAUUG----CCAUGGUGGCAAAAACCA- ...(((----(((.((((((....((((..(.......)..)))).......(((((....)))))..)))))))))).---))..(((----((.....)))))......- ( -31.30) >DroAna_CAF1 18076 87 - 1 UGCAC----GCUGAUUGCCCCAAAGCUGGUCAAAAUCUGAACGA----CAAACAAGUACAAAUUUGUUGAAAUGCA------------G----UCAUGACGGCAAAAACCG- (((.(----(((((((((.(((....)))............(((----((((..........)))))))....)))------------)----))).).))))).......- ( -19.00) >consensus UGAGCG___GCUGAUUGCCCCAAAGCUGGACAAAAUCAGAACGGCAAACAAACAAAUAUAAAUUUGCUGGGCAGCAGCA___GCAAUAG____CCAUGGUGGCAAAAACCG_ ..............((((((((..((((((.....))....(((((((..............)))))))..)))).((....))............))).)))))....... (-16.25 = -17.26 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:30 2006