
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,870,430 – 7,870,686 |
| Length | 256 |
| Max. P | 0.998129 |

| Location | 7,870,430 – 7,870,546 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.94 |
| Mean single sequence MFE | -41.04 |
| Consensus MFE | -37.14 |
| Energy contribution | -37.54 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7870430 116 + 27905053 GUGAGUGUACCACCACUGGCCUGCAAUCAUCCGGACUGGCUCCGGUUGACAUAUGUGGUACUGUGACCGUAUAUGUUCUGCCAUCUGUUAACCCGGAGACGGGCCCAUGAGCCAAA---- ((((.(((((((....)))..)))).))))......((((((.(((.((((((((((((......))))))))))))..))).........((((....)))).....))))))..---- ( -41.60) >DroSec_CAF1 2244 117 + 1 GUGAGUGUACCACCACUGGCCUGCAAUCAUCCGGACUGGCUCCGGUUGACAUAUGUGGUACUGUGACCGUAUAUGUUCUGCCAUCUGUUAGCCCGGAGACGGGCCCAUGAGCCAAAA--- ((((.(((((((....)))..)))).))))......((((((.(((.((((((((((((......))))))))))))..)))........(((((....)))))....))))))...--- ( -45.10) >DroSim_CAF1 2252 117 + 1 GUGAGUGUACCACCACUGGCCUGCAAUCAUCCGGACUGGCUCCGGUUGACAUAUGUGGUACUGUGACCGUAUAUGUUCUGCCAUCUGUUAACCCGGAGACGGGCCCAUGAGCCAAAA--- ((((.(((((((....)))..)))).))))......((((((.(((.((((((((((((......))))))))))))..))).........((((....)))).....))))))...--- ( -41.60) >DroEre_CAF1 2304 116 + 1 GCGAGUGUACCACCACUGGCCUGCAAUCAUCCGGACUGGCUCCGGUUGACAUAUGUGGUACUGUGACCGUAUAUGUUCUGCCAUCUGUUAACCCGGAGACGGGACCAUGAACCAAA---- ((.((((......)))).))..(((.(((.(((((.....))))).)))((((((((((......))))))))))...)))..........((((....)))).............---- ( -38.00) >DroYak_CAF1 2274 120 + 1 GUGAGUGUACCACCACUGGCCUGCAAUCAUCCGGACUGGCUCCGGUUGACAUAUGUGGCACUGUGACCGUAUAUGUUCUGCCAUCUGUUAACCCGGAGACGGGCCCAUGAACGAGAAGGG (((.(((...)))))).((((((...((.(((((..((((...(((.((((((((((((.....).)))))))))))..)))....))))..)))))))))))))............... ( -38.90) >consensus GUGAGUGUACCACCACUGGCCUGCAAUCAUCCGGACUGGCUCCGGUUGACAUAUGUGGUACUGUGACCGUAUAUGUUCUGCCAUCUGUUAACCCGGAGACGGGCCCAUGAGCCAAAA___ ...((((......))))((((((...((.(((((..((((...(((.((((((((((((......))))))))))))..)))....))))..)))))))))))))............... (-37.14 = -37.54 + 0.40)



| Location | 7,870,470 – 7,870,573 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.40 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -27.86 |
| Energy contribution | -28.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7870470 103 + 27905053 UCCGGUUGACAUAUGUGGUACUGUGACCGUAUAUGUUCUGCCAUCUGUUAACCCGGAGACGGGCCCAUGAGCCAAA-------AAGGGGACCAAAAGGGAACGGGGCGCA---------- .(((((.((((((((((((......))))))))))))..)))..(((((..((((....))))(((.((..((...-------....))..))...))))))))))....---------- ( -36.10) >DroSec_CAF1 2284 104 + 1 UCCGGUUGACAUAUGUGGUACUGUGACCGUAUAUGUUCUGCCAUCUGUUAGCCCGGAGACGGGCCCAUGAGCCAAAA------AAGGGGACCAAAAGGGUAAGGGGCGCA---------- ...(((.((((((((((((......))))))))))))..)))........(((((....)))((((....(((....------..(....)......)))...)))))).---------- ( -38.20) >DroSim_CAF1 2292 104 + 1 UCCGGUUGACAUAUGUGGUACUGUGACCGUAUAUGUUCUGCCAUCUGUUAACCCGGAGACGGGCCCAUGAGCCAAAA------AAGGGGACCAAAAGGGUAAGGGGCGCA---------- ...(((.((((((((((((......))))))))))))..)))..........(((....)))((((....(((....------..(....)......)))...))))...---------- ( -36.40) >DroEre_CAF1 2344 112 + 1 UCCGGUUGACAUAUGUGGUACUGUGACCGUAUAUGUUCUGCCAUCUGUUAACCCGGAGACGGGACCAUGAACCAAA--------AAAGGGCUAAAAGGGAACUGGGUGCAGAGGUUCGGA (((((..((((((((((((......))))))))))))..(((.(((((...((((....))))(((.....((...--------...))......((....)).)))))))))))))))) ( -39.40) >DroYak_CAF1 2314 110 + 1 UCCGGUUGACAUAUGUGGCACUGUGACCGUAUAUGUUCUGCCAUCUGUUAACCCGGAGACGGGCCCAUGAACGAGAAGGGGGUAAAGGGGCUAAAAGGAAGCGGGGCGCA---------- ((((((.((((((((((((.....).)))))))))))..))).(((.((..((((....))))(((...........)))....)).)))......))).(((...))).---------- ( -33.40) >consensus UCCGGUUGACAUAUGUGGUACUGUGACCGUAUAUGUUCUGCCAUCUGUUAACCCGGAGACGGGCCCAUGAGCCAAAA______AAGGGGACCAAAAGGGAACGGGGCGCA__________ ...(((.((((((((((((......))))))))))))..)))..........(((....)))((((.....((..............)).((....)).....))))............. (-27.86 = -28.06 + 0.20)

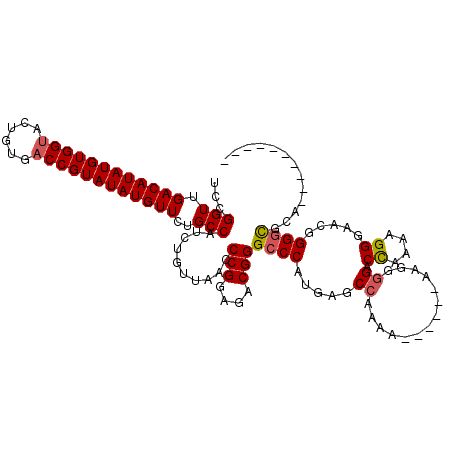

| Location | 7,870,573 – 7,870,686 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.73 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -27.54 |
| Energy contribution | -27.58 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7870573 113 + 27905053 ------GAGGUUCAGCUGUCACUCAGAUCUCUCCGCUUUGGUUUAACAUUUCCCUUU-UUUAGCUUAUCCCAGUGACCUCACUGUGGGGAUUUUGCUUUUUGGGGAUUUUAUUGACUCUG ------(((((....(((.....))))))))........((((.......(((((..-...(((..(((((((((....))))...)))))...)))....))))).......))))... ( -28.54) >DroSec_CAF1 2388 114 + 1 ------GAGGUUCAGCUGUCACUCAGAUCUCUCCGCUUUGGUUUAACAUUUCCCUUUUUUUAGCUUGUCCCAGUGACCUCACUGUGGGGAUUUUGCUUUUUGGGGAUUUUAUUGACUCUG ------(((((....(((.....))))))))........((((.......(((((......(((..(((((((((....))))...)))))...)))....))))).......))))... ( -28.24) >DroSim_CAF1 2396 114 + 1 ------GAGGUUCAGCUGUCACUCAGAUCUCUCCGCUUUGGUUUAACAUUUCCCUUUUUUUAGCUUAUCCCAGUGACCUCACUGUGGGGAUUUUGCUUUUUGGGGAUUUUAUUGACUCUG ------(((((....(((.....))))))))........((((.......(((((......(((..(((((((((....))))...)))))...)))....))))).......))))... ( -27.94) >DroEre_CAF1 2456 119 + 1 GGCUCAGAGGUUCAGCUGUCACUCAGAUCUCUCCGCUUUGGUUUAACAUUUCCCAUU-UUUAGCUUAUCCCAGUGACCUCACUGUGGGGAUUUUGCUUUUUGGGGAUUUUAUUGACUCUG (((..((((((....(((.....)))))))))..)))..((((......((((((..-...(((..(((((((((....))))...)))))...)))...)))))).......))))... ( -32.02) >DroYak_CAF1 2424 113 + 1 ------GAGGUUCAGCUGUCACUCAGAUCUCUCCGCUUUGGUUUAACAUUUCCCUUU-UUUAGCUUAUCCCAGUGACCUCACUGUGGGGAUUUUGCUUUUUGGGGAUUUUAUUGACUCUG ------(((((....(((.....))))))))........((((.......(((((..-...(((..(((((((((....))))...)))))...)))....))))).......))))... ( -28.54) >consensus ______GAGGUUCAGCUGUCACUCAGAUCUCUCCGCUUUGGUUUAACAUUUCCCUUU_UUUAGCUUAUCCCAGUGACCUCACUGUGGGGAUUUUGCUUUUUGGGGAUUUUAUUGACUCUG ......(((((....(((.....))))))))........((((.......(((((......(((..(((((((((....))))...)))))...)))....))))).......))))... (-27.54 = -27.58 + 0.04)



| Location | 7,870,573 – 7,870,686 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.73 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -25.04 |
| Energy contribution | -25.24 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.998129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7870573 113 - 27905053 CAGAGUCAAUAAAAUCCCCAAAAAGCAAAAUCCCCACAGUGAGGUCACUGGGAUAAGCUAAA-AAAGGGAAAUGUUAAACCAAAGCGGAGAGAUCUGAGUGACAGCUGAACCUC------ (((.((((......((((.....(((...(((((...((((....)))))))))..)))...-...))))...............((((....))))..))))..)))......------ ( -26.10) >DroSec_CAF1 2388 114 - 1 CAGAGUCAAUAAAAUCCCCAAAAAGCAAAAUCCCCACAGUGAGGUCACUGGGACAAGCUAAAAAAAGGGAAAUGUUAAACCAAAGCGGAGAGAUCUGAGUGACAGCUGAACCUC------ (((.((((......((((.....(((....((((...((((....))))))))...))).......))))...............((((....))))..))))..)))......------ ( -24.00) >DroSim_CAF1 2396 114 - 1 CAGAGUCAAUAAAAUCCCCAAAAAGCAAAAUCCCCACAGUGAGGUCACUGGGAUAAGCUAAAAAAAGGGAAAUGUUAAACCAAAGCGGAGAGAUCUGAGUGACAGCUGAACCUC------ (((.((((......((((.....(((...(((((...((((....)))))))))..))).......))))...............((((....))))..))))..)))......------ ( -25.50) >DroEre_CAF1 2456 119 - 1 CAGAGUCAAUAAAAUCCCCAAAAAGCAAAAUCCCCACAGUGAGGUCACUGGGAUAAGCUAAA-AAUGGGAAAUGUUAAACCAAAGCGGAGAGAUCUGAGUGACAGCUGAACCUCUGAGCC ((((((((......((((.....(((...(((((...((((....)))))))))..)))...-...))))..(((((........((((....))))..)))))..)))..))))).... ( -29.00) >DroYak_CAF1 2424 113 - 1 CAGAGUCAAUAAAAUCCCCAAAAAGCAAAAUCCCCACAGUGAGGUCACUGGGAUAAGCUAAA-AAAGGGAAAUGUUAAACCAAAGCGGAGAGAUCUGAGUGACAGCUGAACCUC------ (((.((((......((((.....(((...(((((...((((....)))))))))..)))...-...))))...............((((....))))..))))..)))......------ ( -26.10) >consensus CAGAGUCAAUAAAAUCCCCAAAAAGCAAAAUCCCCACAGUGAGGUCACUGGGAUAAGCUAAA_AAAGGGAAAUGUUAAACCAAAGCGGAGAGAUCUGAGUGACAGCUGAACCUC______ (((.((((......((((.....(((...(((((...((((....)))))))))..))).......))))...............((((....))))..))))..)))............ (-25.04 = -25.24 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:18 2006