
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,863,870 – 7,864,011 |
| Length | 141 |
| Max. P | 0.992821 |

| Location | 7,863,870 – 7,863,976 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 92.45 |
| Mean single sequence MFE | -38.24 |
| Consensus MFE | -35.70 |
| Energy contribution | -35.22 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7863870 106 + 27905053 AUGCAUUUUCAGCUGCAUGUGACUGGGACGAUGGGUUGCCAAAA-UG------------GGCUGGUGGCUAGUGGAUAUGAGGUCAAAGACCCUUAUACCCUGGCUGAGAGCUAUCUCA ........((((((.((((..(((.(.....).)))..)....)-))------------)))))).((((((.((.(((((((........)))))))))))))))((((....)))). ( -34.10) >DroSec_CAF1 24706 106 + 1 AUGCAUUUUCAGCUGCAUGUGACUGGGACGAUGGGUUGCCAAAA-UG------------GGCUGGUGGCCAGUGGAUAUGAGGUCAAAGACCCUUAUACCCUGGCUGAGAGCUAUCUCA ........((((((.((((..(((.(.....).)))..)....)-))------------)))))).((((((.((.(((((((........)))))))))))))))((((....)))). ( -36.40) >DroSim_CAF1 25005 106 + 1 AUGCAUUUUCAGCUGCAUGUGACUGGGACGAUGGGUUGCCAAAA-UG------------GGCUGGUGGCCAGUGGAUAUGAGGUCAAAGACCCUUAUACCCUGGCUGAGAGCUAUCUCA ........((((((.((((..(((.(.....).)))..)....)-))------------)))))).((((((.((.(((((((........)))))))))))))))((((....)))). ( -36.40) >DroEre_CAF1 24868 106 + 1 AUGCAUUUUCAGCUGCAUGUGACUGGGACGAUGGGUGGCCAAAA-UG------------GGCUGGCGGCCAGUGGAUAUGAGGUCAAAGACCCUUAUACCCUGGCUGAGAGCUAUCUCU (((((........)))))......((((...(((....)))...-..------------((((..(((((((.((.(((((((........))))))))))))))))..)))).)))). ( -37.50) >DroYak_CAF1 25226 119 + 1 GUGCAGUUUCAGCUGCAUGUGACUGGGACGAUGGGUGGCCAAAAAUGGUGGUGCCUGGUGGCUGGUGGCCAGUGGAUAUGAGGUCAACGACCCUUAUACCCUGGCUGAGAGCUAUCUCU (((((((....)))))))..............(((((((((....))))..)))))(((((((..(((((((.((.(((((((........))))))))))))))))..)))))))... ( -46.80) >consensus AUGCAUUUUCAGCUGCAUGUGACUGGGACGAUGGGUUGCCAAAA_UG____________GGCUGGUGGCCAGUGGAUAUGAGGUCAAAGACCCUUAUACCCUGGCUGAGAGCUAUCUCA (((((........)))))......((((...(((....)))..................((((..(((((((.((.(((((((........))))))))))))))))..)))).)))). (-35.70 = -35.22 + -0.48)

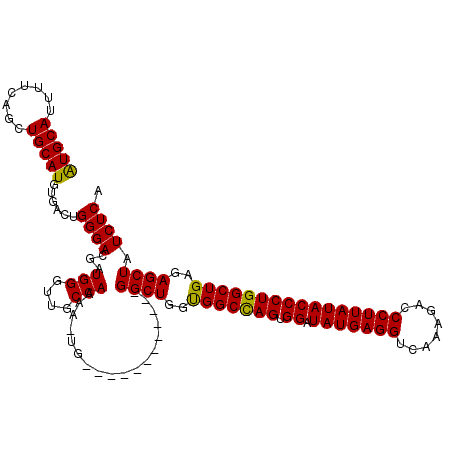

| Location | 7,863,870 – 7,863,976 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.45 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -25.64 |
| Energy contribution | -26.04 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7863870 106 - 27905053 UGAGAUAGCUCUCAGCCAGGGUAUAAGGGUCUUUGACCUCAUAUCCACUAGCCACCAGCC------------CA-UUUUGGCAACCCAUCGUCCCAGUCACAUGCAGCUGAAAAUGCAU (((((....)))))((((((((((.(((........))).))))))....((.....)).------------..-...))))...................(((((........))))) ( -25.40) >DroSec_CAF1 24706 106 - 1 UGAGAUAGCUCUCAGCCAGGGUAUAAGGGUCUUUGACCUCAUAUCCACUGGCCACCAGCC------------CA-UUUUGGCAACCCAUCGUCCCAGUCACAUGCAGCUGAAAAUGCAU (((((....)))))(((((((.....(((((...))))).....)).))))).....(((------------..-....)))...................(((((........))))) ( -28.50) >DroSim_CAF1 25005 106 - 1 UGAGAUAGCUCUCAGCCAGGGUAUAAGGGUCUUUGACCUCAUAUCCACUGGCCACCAGCC------------CA-UUUUGGCAACCCAUCGUCCCAGUCACAUGCAGCUGAAAAUGCAU (((((....)))))(((((((.....(((((...))))).....)).))))).....(((------------..-....)))...................(((((........))))) ( -28.50) >DroEre_CAF1 24868 106 - 1 AGAGAUAGCUCUCAGCCAGGGUAUAAGGGUCUUUGACCUCAUAUCCACUGGCCGCCAGCC------------CA-UUUUGGCCACCCAUCGUCCCAGUCACAUGCAGCUGAAAAUGCAU .((((....)))).(((((((.....(((((...))))).....)).))))).(((((..------------..-..)))))...................(((((........))))) ( -29.30) >DroYak_CAF1 25226 119 - 1 AGAGAUAGCUCUCAGCCAGGGUAUAAGGGUCGUUGACCUCAUAUCCACUGGCCACCAGCCACCAGGCACCACCAUUUUUGGCCACCCAUCGUCCCAGUCACAUGCAGCUGAAACUGCAC .((((....)))).(((.((((((.(((........))).))))))..((((.....))))...)))....(((....))).....................(((((......))))). ( -32.20) >consensus UGAGAUAGCUCUCAGCCAGGGUAUAAGGGUCUUUGACCUCAUAUCCACUGGCCACCAGCC____________CA_UUUUGGCAACCCAUCGUCCCAGUCACAUGCAGCUGAAAAUGCAU .((((....)))).((((((((((.(((........))).))))))...(((.....)))..................))))...................(((((........))))) (-25.64 = -26.04 + 0.40)

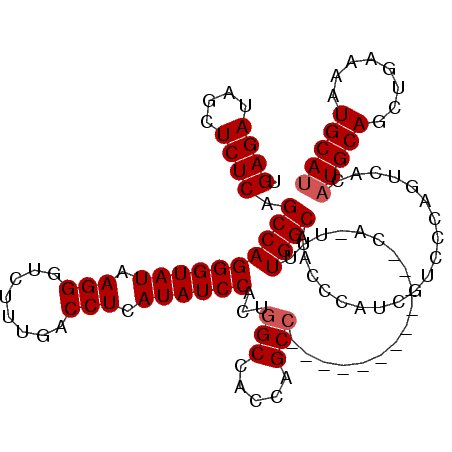

| Location | 7,863,910 – 7,864,011 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -35.14 |
| Consensus MFE | -26.70 |
| Energy contribution | -26.38 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7863910 101 + 27905053 AAAA-UG------------GGCUGGUGGCUAGUGGAUAUGAGGUCAAAGACCCUUAUACCCUGGCUGAGAGCUAUCUCAAUUUGUUUUGGAGUGGCGAGUGUUUUCGUGUUUAU----- ....-..------------((((..(((((((.((.(((((((........))))))))))))))))..))))((..(((......)))..)).(((((....)))))......----- ( -30.00) >DroSec_CAF1 24746 85 + 1 AAAA-UG------------GGCUGGUGGCCAGUGGAUAUGAGGUCAAAGACCCUUAUACCCUGGCUGAGAGCUAUCUCAAUUUGUUUUGA----------------GAACUUGU----- ....-..------------((((..(((((((.((.(((((((........))))))))))))))))..)))).((((((......))))----------------))......----- ( -32.90) >DroSim_CAF1 25045 101 + 1 AAAA-UG------------GGCUGGUGGCCAGUGGAUAUGAGGUCAAAGACCCUUAUACCCUGGCUGAGAGCUAUCUCAAUUUGUUUUGGAGUGGUGAGUGUUUUCGUGUUUAU----- ...(-((------------(((....((((((.((.(((((((........)))))))))))))))((((((...((((((((......))))..)))).))))))..))))))----- ( -30.90) >DroEre_CAF1 24908 106 + 1 AAAA-UG------------GGCUGGCGGCCAGUGGAUAUGAGGUCAAAGACCCUUAUACCCUGGCUGAGAGCUAUCUCUACUUGUUUUGGAGUGGCGAGUGUUUUCGUGUUUAUAUACA ....-..------------((((..(((((((.((.(((((((........))))))))))))))))..)))).((.((((((......)))))).))........((((....)))). ( -37.60) >DroYak_CAF1 25266 119 + 1 AAAAAUGGUGGUGCCUGGUGGCUGGUGGCCAGUGGAUAUGAGGUCAACGACCCUUAUACCCUGGCUGAGAGCUAUCUCUAUUUGUUUUGGAGUGGAGAGUGUUCUCGUGUUUAUAUACA ............(((....)))....((((((.((.(((((((........)))))))))))))))((((((..(((((((((......)))))))))..))))))((((....)))). ( -44.30) >consensus AAAA_UG____________GGCUGGUGGCCAGUGGAUAUGAGGUCAAAGACCCUUAUACCCUGGCUGAGAGCUAUCUCAAUUUGUUUUGGAGUGGCGAGUGUUUUCGUGUUUAU_____ ...................((((..(((((((.((.(((((((........))))))))))))))))..)))).............................................. (-26.70 = -26.38 + -0.32)



| Location | 7,863,910 – 7,864,011 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -22.26 |
| Consensus MFE | -18.60 |
| Energy contribution | -18.80 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7863910 101 - 27905053 -----AUAAACACGAAAACACUCGCCACUCCAAAACAAAUUGAGAUAGCUCUCAGCCAGGGUAUAAGGGUCUUUGACCUCAUAUCCACUAGCCACCAGCC------------CA-UUUU -----..................((..............((((((....))))))...((((((.(((........))).))))))....))........------------..-.... ( -16.50) >DroSec_CAF1 24746 85 - 1 -----ACAAGUUC----------------UCAAAACAAAUUGAGAUAGCUCUCAGCCAGGGUAUAAGGGUCUUUGACCUCAUAUCCACUGGCCACCAGCC------------CA-UUUU -----......((----------------((((......))))))..(((....(((((((.....(((((...))))).....)).)))))....))).------------..-.... ( -24.10) >DroSim_CAF1 25045 101 - 1 -----AUAAACACGAAAACACUCACCACUCCAAAACAAAUUGAGAUAGCUCUCAGCCAGGGUAUAAGGGUCUUUGACCUCAUAUCCACUGGCCACCAGCC------------CA-UUUU -----...................................(((((....)))))(((((((.....(((((...))))).....)).)))))........------------..-.... ( -20.70) >DroEre_CAF1 24908 106 - 1 UGUAUAUAAACACGAAAACACUCGCCACUCCAAAACAAGUAGAGAUAGCUCUCAGCCAGGGUAUAAGGGUCUUUGACCUCAUAUCCACUGGCCGCCAGCC------------CA-UUUU ....................(((...(((........))).)))...(((..(.(((((((.....(((((...))))).....)).))))).)..))).------------..-.... ( -22.40) >DroYak_CAF1 25266 119 - 1 UGUAUAUAAACACGAGAACACUCUCCACUCCAAAACAAAUAGAGAUAGCUCUCAGCCAGGGUAUAAGGGUCGUUGACCUCAUAUCCACUGGCCACCAGCCACCAGGCACCACCAUUUUU .............((((....))))................((((....)))).(((.((((((.(((........))).))))))..((((.....))))...)))............ ( -27.60) >consensus _____AUAAACACGAAAACACUCGCCACUCCAAAACAAAUUGAGAUAGCUCUCAGCCAGGGUAUAAGGGUCUUUGACCUCAUAUCCACUGGCCACCAGCC____________CA_UUUU .........................................((((....)))).(((((((.....(((((...))))).....)).)))))........................... (-18.60 = -18.80 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:08 2006