
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
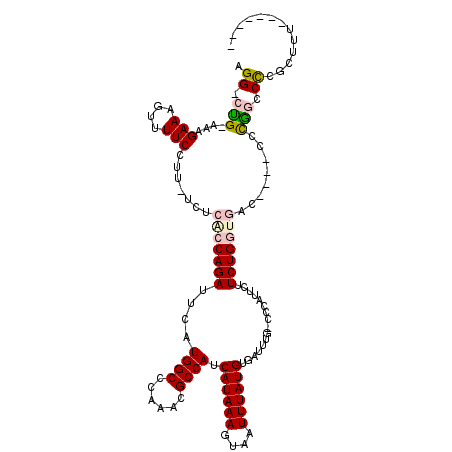
| Location | 7,858,019 – 7,858,126 |
| Length | 107 |
| Max. P | 0.762809 |

| Location | 7,858,019 – 7,858,126 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.38 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -15.80 |
| Energy contribution | -17.72 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7858019 107 + 27905053 AGG-CUG-AAAGAAAGUUUUCCUUCUCUCACCAGAUUCAUGGCCCAAACGCCAUCAUAAAGUAAUUUAUGUGAUUUG-ACCAUUCUUCUGGUGACCAC-CCCAGUCCGGCC--------- .((-(((-..((((........)))).((((((((...((((..((((...((.((((((....)))))))).))))-.))))...))))))))....-.......)))))--------- ( -30.50) >DroPse_CAF1 20907 112 + 1 AGG-CCGGACCGAAAGUUUUCCU------ACCAGAUUCCUGGCUGGGAUGCCAUCAUAAAGUAAUUUAUGUGAUUUGUCCCAUUCUUCUGCGGAGAGGCCCCG-CCUCGUUUUUCUCCGC ...-..((((.((..((..((((------(((((....)))).))))).))((.((((((....)))))))).)).)))).........((((((((((...)-))))(.....)))))) ( -33.90) >DroEre_CAF1 19106 106 + 1 AGG-CUG-AAAGAAAGUUUUCCCUCUCUCACCAGAUUCAUGGCCCAAGCGCCAUCAUAAAGUAAUUUAUGUGAUUUG-CCCAUUCUUCUGGUGAC----CCCGGUCCCGCCAU------- .((-(.(-((((....)))))((....((((((((...((((..((((...((.((((((....)))))))).))))-.))))...)))))))).----...))....)))..------- ( -28.90) >DroYak_CAF1 19363 106 + 1 AGG-CUG-AAAGAAAGUUUUCCUUCUCUCACCAGAUUCAUGGCCCAAACGCCAUCAUAAAGUAAUUUAUGUGAUUUG-CCCAUUCUUCUGGUGAC----CCUGGUCCGGCGGC------- ..(-(((-..((((........)))).((((((((...((((..((((...((.((((((....)))))))).))))-.))))...)))))))).----.......))))...------- ( -27.90) >DroAna_CAF1 21183 106 + 1 GGGCCUG-AAAGAAACUUUUCCUU-ACACGCCAGAUUCAUGGCCCGGUCGCCAUCAUAAAGUAAUUUAUGUGAUUUG-CCCAUUCUUCUGGUGAC----CCCAGCCUGCCUCC------- ((((..(-((((....)))))...-...(((((((...((((.(..(((((....(((((....))))))))))..)-.))))...)))))))..----....))))......------- ( -29.20) >DroPer_CAF1 20923 105 + 1 AGG-CCGGACCGAAAGUUUUCCU------ACCAGAUUCCUGGCUGGGAUGCCAUCAUAAAGUAAUUUAUGUGAUUUGUCCCAUUCUUCUGCGGAGAGGCCCCG-CCUCGUUUU------- .((-((((((.((..((..((((------(((((....)))).))))).))((.((((((....)))))))).)).))))...((((.....))))))))...-.........------- ( -32.50) >consensus AGG_CUG_AAAGAAAGUUUUCCUU_UCUCACCAGAUUCAUGGCCCAAACGCCAUCAUAAAGUAAUUUAUGUGAUUUG_CCCAUUCUUCUGGUGAC____CCCGGCCCCGCUUU_______ .((.(((....(((....))).......(((((((....((((......)))).((((((....))))))................)))))))........))).))............. (-15.80 = -17.72 + 1.92)

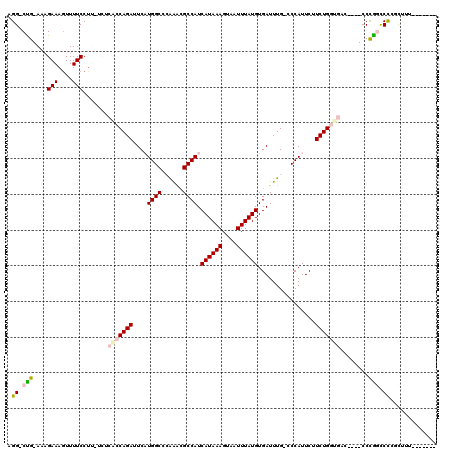
| Location | 7,858,019 – 7,858,126 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.38 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -14.94 |
| Energy contribution | -14.42 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7858019 107 - 27905053 ---------GGCCGGACUGGG-GUGGUCACCAGAAGAAUGGU-CAAAUCACAUAAAUUACUUUAUGAUGGCGUUUGGGCCAUGAAUCUGGUGAGAGAAGGAAAACUUUCUUU-CAG-CCU ---------(((..((((...-..))))((((((...(((((-(((((((((((((....)))))).))..)))).))))))...))))))(((((((((....))))))))-).)-)). ( -37.50) >DroPse_CAF1 20907 112 - 1 GCGGAGAAAAACGAGG-CGGGGCCUCUCCGCAGAAGAAUGGGACAAAUCACAUAAAUUACUUUAUGAUGGCAUCCCAGCCAGGAAUCUGGU------AGGAAAACUUUCGGUCCGG-CCU (((((((.......((-(...)))))))))).......(((((....(((((((((....)))))).)))..)))))(((.(((..((((.------((.....)).)))))))))-).. ( -34.81) >DroEre_CAF1 19106 106 - 1 -------AUGGCGGGACCGGG----GUCACCAGAAGAAUGGG-CAAAUCACAUAAAUUACUUUAUGAUGGCGCUUGGGCCAUGAAUCUGGUGAGAGAGGGAAAACUUUCUUU-CAG-CCU -------..(((..(((....----)))((((((...((((.-(...(((((((((....)))))).)))......).))))...))))))(((((((((....))))))))-).)-)). ( -36.00) >DroYak_CAF1 19363 106 - 1 -------GCCGCCGGACCAGG----GUCACCAGAAGAAUGGG-CAAAUCACAUAAAUUACUUUAUGAUGGCGUUUGGGCCAUGAAUCUGGUGAGAGAAGGAAAACUUUCUUU-CAG-CCU -------......(((((((.----.((.(((......))).-.......((((((....))))))(((((......)))))))..)))))(((((((((....))))))))-)..-)). ( -32.80) >DroAna_CAF1 21183 106 - 1 -------GGAGGCAGGCUGGG----GUCACCAGAAGAAUGGG-CAAAUCACAUAAAUUACUUUAUGAUGGCGACCGGGCCAUGAAUCUGGCGUGU-AAGGAAAAGUUUCUUU-CAGGCCC -------(((..((((((.((----(((((((......))).-.......((((((....)))))).))))..)).)))).))..)))(((.((.-((((((....))))))-)).))). ( -34.00) >DroPer_CAF1 20923 105 - 1 -------AAAACGAGG-CGGGGCCUCUCCGCAGAAGAAUGGGACAAAUCACAUAAAUUACUUUAUGAUGGCAUCCCAGCCAGGAAUCUGGU------AGGAAAACUUUCGGUCCGG-CCU -------......(((-(.((((((((....))).((((((((....(((((((((....)))))).)))..)))))(((((....)))))------.........)))))))).)-))) ( -34.30) >consensus _______AAAGCCAGACCGGG____GUCACCAGAAGAAUGGG_CAAAUCACAUAAAUUACUUUAUGAUGGCGUCCGGGCCAUGAAUCUGGUGAGA_AAGGAAAACUUUCUUU_CAG_CCU ...............((((((.....((((((......))).........((((((....)))))).((((......))))))).))))))............................. (-14.94 = -14.42 + -0.52)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:01 2006