
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,857,721 – 7,857,826 |
| Length | 105 |
| Max. P | 0.543330 |

| Location | 7,857,721 – 7,857,826 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 83.22 |
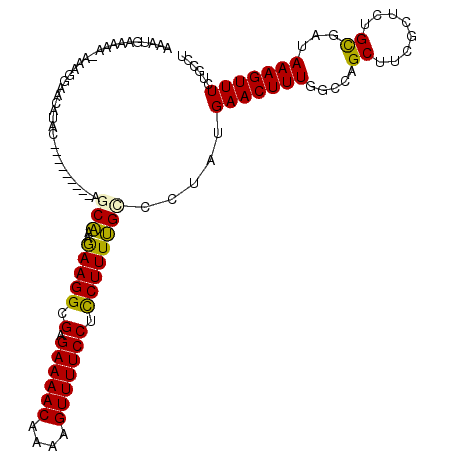
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -15.82 |
| Energy contribution | -15.30 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
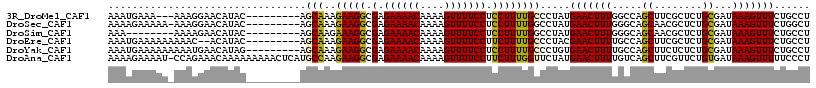
>3R_DroMel_CAF1 7857721 105 - 27905053 AAAUGAAA---AAAGGAACAUAC---------AGCAAAGAAGGCGAGAAAACAAAAGUUUUCCUCCUUUUGCCCUAUGAACUUUGGCCAGCUUCGCUCUGCGAUAAAGUUUCUGCCU ........---((((...((((.---------.(((..(((((.(.((((((....))))))).))))))))..))))..))))(((.((((((((...))))...))))...))). ( -26.10) >DroSec_CAF1 18645 107 - 1 AAAAGAAAAA-AAAGGAACAUAC---------AGCAAAGAAGGCGAGAAAACAAAAGUUUUCCUCCUUUUGGCCUAUGAACUUUGGGCAGCAACGCUCUGCGAUAAAGUUUCUGGCU ..........-..((((((....---------.((..((((((.(.((((((....))))))).)))))).(((((.......))))).))..(((...))).....)))))).... ( -28.20) >DroSim_CAF1 18929 100 - 1 AAA--------AAAAGAACAUAC---------AGCAAGAAAGGCGAGAAAACAAAAGUUUUCCUCCUUUUGGCCUAUGAACUUUGGGCAGCAACGCUCUGCGAUAAAGUUUCUGCCU ...--------.((((..((((.---------.((..((((((.(.((((((....))))))).)))))).)).))))..))))((((((.(((.............))).)))))) ( -26.62) >DroEre_CAF1 18811 106 - 1 AAAUGAAAAAAAAAC--ACAUAC---------AGCAAAGAAGGCGAGAAAACAAAAGUUUUCCUUCUUUUGCCCUACGAACUUUUGCCAGCUUCGCUCUGCGAUAAAGUUUCUGCCU ...............--......---------.((((((((((....(((((....)))))))))).))))).....((((((((((.(((...)))..))))..))))))...... ( -18.40) >DroYak_CAF1 19069 108 - 1 AAAUGAAAAAAAAAUGAACAUAG---------AGCAAAGAAGGCGAGAAAACAAAAGUUUUCCUCCUUUUGCCCUGUGAACUUUUGCCAGCUUCUCUCUGCGAUAAAGUUUCUGCCU ..................(((((---------.(((..(((((.(.((((((....))))))).)))))))).)))))((((((.....((........))...))))))....... ( -24.50) >DroAna_CAF1 20896 116 - 1 AAAAGAAAAU-CCAGAAACAAAAAAAAACUCAUGCCAAGAAGGCGAGAAAACAAAAGUUUUCCUUCUUUGGUUCUAUGAACUUUUGUCAGCUUCGUUCUGUGAUAAAGUUUUUCCCU ........((-((((((................(((.....)))(((...((((((((((.((......))......))))))))))...)))..))))).)))............. ( -22.30) >consensus AAAUGAAAAA_AAAGGAACAUAC_________AGCAAAGAAGGCGAGAAAACAAAAGUUUUCCUCCUUUUGCCCUAUGAACUUUGGCCAGCUUCGCUCUGCGAUAAAGUUUCUGCCU .................................(((..(((((.(.((((((....))))))).)))))))).....(((((((.....((........))...)))))))...... (-15.82 = -15.30 + -0.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:59 2006