
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,824,848 – 7,824,940 |
| Length | 92 |
| Max. P | 0.533859 |

| Location | 7,824,848 – 7,824,940 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 65.90 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -6.30 |
| Energy contribution | -6.88 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.27 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7824848 92 - 27905053 GAGGCCAGCGCAAUUGCAGCGCAGCGGCAACAUUGU-UGAGCAACAUUUGUGCUAAACUAU-----GGAACAAAGA-AAUAACUAAAGAA--GUAGAAG----CU------ ..(((.(((((((.((..((.((((((.....))))-)).))..)).)))))))...((((-----.......((.-.....))......--))))..)----))------ ( -20.12) >DroSec_CAF1 13789 87 - 1 GAGGCCAGCGCAAUUGCAGCGCAGCGGCAACAUUGU-UGAGCAACAUUUGUCCUAAACUAU-----GGGACAAAGA-AAAAACUACGGAA--GUAA--------------- ...(((.((((.......))))...))).....(((-(....))))(((((((((.....)-----))))))))..-......(((....--))).--------------- ( -25.30) >DroSim_CAF1 15710 87 - 1 GAGGCCAGCGCAAUUGCAGCGCAGCGGCAACAUUGU-UGAGCAACAUUUGUCCUAAACUAU-----GGGACAAAGA-AAAAACUAAGGAA--GUAA--------------- ....((.((((.......)))).((..((((...))-)).))....(((((((((.....)-----))))))))..-.........))..--....--------------- ( -22.80) >DroEre_CAF1 15997 80 - 1 GAGGCCAGCGCAAUUGCAGCGCGGCGGCAGCAUUGU-UGAGCAACAUUUUGG----------------------CUAAGCAACUAACGAA--AUAAAAAAACACU------ ..(((((((((.......)))).((..((((...))-)).)).......)))----------------------))..............--.............------ ( -18.20) >DroYak_CAF1 16228 89 - 1 GAGGCUAGCGCAAUUGCAGC-CAGCGGCAACAUUGU-UGGGCAACAUUUGGU-GGAACAAU-----GGAGCACACAGAAUAACUAAAGAA--AUCAAAA------------ ((((((.(((....))))))-).(.(((..((((((-(..((........))-..))))))-----)..)).).)...............--.))....------------ ( -20.10) >DroPer_CAF1 14938 105 - 1 GAGGCGCAGGCAAUUGCAUUGCGCCAGCAACAUUUUCUGGGUAACGAA-GUGUUGAAAGAUACGAAGACGUAAUGA-AACAACUUAGAAGAUGCAGAAC----CGAAGACG ..(((((((((....)).))))))).....((((((((((((......-(((((....)))))......((.....-.)).))))))))))))......----........ ( -32.40) >consensus GAGGCCAGCGCAAUUGCAGCGCAGCGGCAACAUUGU_UGAGCAACAUUUGUGCUAAACUAU_____GGAACAAAGA_AAAAACUAAGGAA__GUAAAA_____________ (..(((.((((.......))))...)))..)................................................................................ ( -6.30 = -6.88 + 0.59)

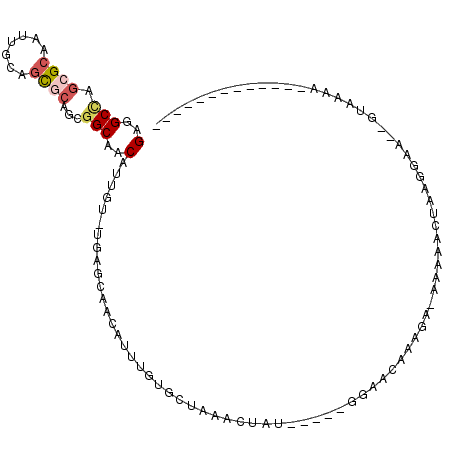
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:44 2006