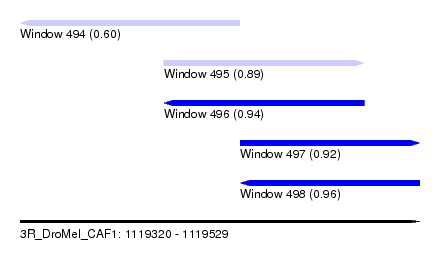
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,119,320 – 1,119,529 |
| Length | 209 |
| Max. P | 0.964390 |

| Location | 1,119,320 – 1,119,435 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 83.87 |
| Mean single sequence MFE | -35.02 |
| Consensus MFE | -28.93 |
| Energy contribution | -29.32 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1119320 115 - 27905053 AACACCGUCCGAAGACUCUGGUGGCUCACCUGUGCCGCCAUUGACUCCGCAUUCGACUCAUUCCGAGGGUCACAUGACCUAUGGGUAAGGACGGAUCUCCGAUCAGCUGUCACCA ....(((((((.((.(..(((((((.(....).)))))))..).)).).............((((.(((((....))))).))))...))))))......(((.....))).... ( -37.50) >DroVir_CAF1 46912 105 - 1 AACACCGUCCGAAGAAUCUGGUGGGUCGCCAGUGCCGCCAUUGACUCCACACUCGACACAUUCCGAGGGUCACAUGACCUAUGGGUAAGGACGGAUU--GGAUCC--UG------ ....((((((........((((((..(....)..))))))...((.(((..((((........))))((((....))))..)))))..))))))...--......--..------ ( -34.10) >DroGri_CAF1 60554 106 - 1 AACACCGUCCGAAGAAUCUGGUGGGUCGCCAGUGCCGCCAUUAACUCCACAUUCGACACAUUCCGAGGGUCACAUGACCUAUGGGUAAGGACGGAUUU-GGAUGA--UG------ ....(((((((..((((.(((..(((.((....)).))).......))).))))..)....((((.(((((....))))).))))...))))))....-......--..------ ( -34.00) >DroWil_CAF1 56051 105 - 1 AACACCAUCCGAAGACUCUGGUGGCUCACCAGUGCCGCCAUUGACUCCCCAUUCAACACAUUCCGAGGGUCACAUGACCUAUGGGUAAGGCAGUACUU-GUAUAC---------U ....(((((.((....)).)))))...((.(((((.(((.((((........)))).....((((.(((((....))))).))))...))).))))).-))....---------. ( -30.40) >DroMoj_CAF1 48002 105 - 1 AACACCGUCCGAAGAAUCUGGUGGAUCACCAGUGCCGCCAUUGACACCGCACUCGACACAUUCCGAGGGUCAUAUGACCUAUGGGUAAGGAUGGAUC--GGCAAC--UG------ ..(((((...........)))))......(((((((((((((...(((.((((((........))))((((....))))..)))))...)))))..)--))).))--))------ ( -32.90) >DroAna_CAF1 50713 102 - 1 AACACCGUCCGAGGACUCUGGUGGCUCACCUGUGCCGCCAUUGACUCCGCACUCCACUCAUUCCGAGGGUCACAUGACCUAUGGGUAAGGACGGAUCUA---UC----------C ....(((((((.(((.(((((((((.(....).)))))))..)).))).)...........((((.(((((....))))).))))...)))))).....---..----------. ( -41.20) >consensus AACACCGUCCGAAGAAUCUGGUGGCUCACCAGUGCCGCCAUUGACUCCGCACUCGACACAUUCCGAGGGUCACAUGACCUAUGGGUAAGGACGGAUCU_GGAUCC__UG______ ....((((((........((((((..........)))))).....................((((.(((((....))))).))))...))))))..................... (-28.93 = -29.32 + 0.39)

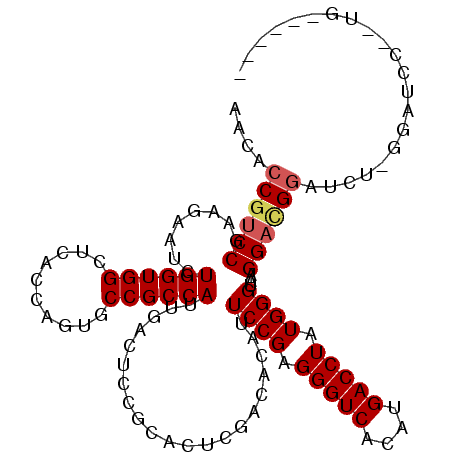
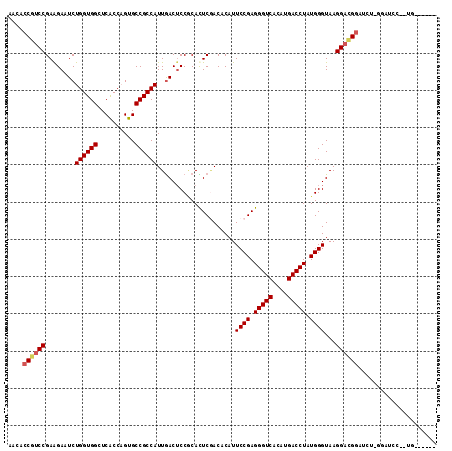
| Location | 1,119,395 – 1,119,500 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.04 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -29.16 |
| Energy contribution | -28.97 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1119395 105 + 27905053 UGGCGGCACAGGUGAGCCACCAGAGUCUUCGGACGGUGUUUUCGAAAAGAGAUUCCCGCCCAGCUCUGAGACCAGAGUAAUGGGCGAGCUAAAUG---------------GUUCGUACAU (((.(((........))).)))..(((....)))((...((((.....))))...))(((((((((((....))))))..)))))(((((....)---------------))))...... ( -40.50) >DroVir_CAF1 46977 105 + 1 UGGCGGCACUGGCGACCCACCAGAUUCUUCGGACGGUGUUUUCGAAAACAAGUUGCCGCCCAGCUCUGAUACCAGAGUGAUGGGCGAGCUAAGUA---------------AUUCAUUUAU (((((((((((.(((..((((....((....)).))))...)))....).)).))))(((((((((((....))))))..)))))..))))....---------------.......... ( -37.00) >DroPse_CAF1 59612 104 + 1 UGGCGGCACUGGCGAGCCACCAGAGUCUUCGGACGGUGUUUUCGAAAACAGAUUCCCGCCCAGCUCUGAGACCAGAGUAAUGGGCGAGCUAUAGA---------------AUU-UUGUGU ((((((..(((.((((.((((...(((....)))))))..))))....)))...))((((((((((((....))))))..)))))).))))....---------------...-...... ( -38.90) >DroGri_CAF1 60620 120 + 1 UGGCGGCACUGGCGACCCACCAGAUUCUUCGGACGGUGUUUUCGAAAAUAGAUUGCCGCCCAGCUCUGAGACCAGAGUGAUGGGUGAGCUAAGUAAUUGCAUAUAAACAUUUUUUUUUUU .((((((((((.(((..((((....((....)).))))...)))....)))..)))))))..((((((....))))))((((.(((.((.((....)))).)))...))))......... ( -35.30) >DroMoj_CAF1 48067 108 + 1 UGGCGGCACUGGUGAUCCACCAGAUUCUUCGGACGGUGUUUUCGAAAAUAGAUUGCCGCCCAGCUCUGAGACCAGAGUGAUGGGCGAGCUGAUUU---G---------AUUUUGAUUUUU .((((.(.((((((...))))))..((....)).).)))).((((((.((((((((((((((((((((....))))))..)))))).)).)))))---)---------.))))))..... ( -41.20) >DroAna_CAF1 50775 92 + 1 UGGCGGCACAGGUGAGCCACCAGAGUCCUCGGACGGUGUUUUCGAAAAUAGAUUCCCGCCCAGCUCUGACACCAGAGUGAUGGGCGAGCUAA---------------------------- (((((((((.((((...))))...(((....))).)))))................((((((((((((....))))))..)))))).)))).---------------------------- ( -35.20) >consensus UGGCGGCACUGGCGAGCCACCAGAGUCUUCGGACGGUGUUUUCGAAAAUAGAUUCCCGCCCAGCUCUGAGACCAGAGUGAUGGGCGAGCUAAAUA_______________AUU_AUUUAU (((((((((((.(((.............)))..)))))))................((((((((((((....))))))..)))))).))))............................. (-29.16 = -28.97 + -0.19)
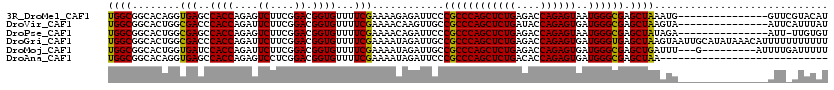
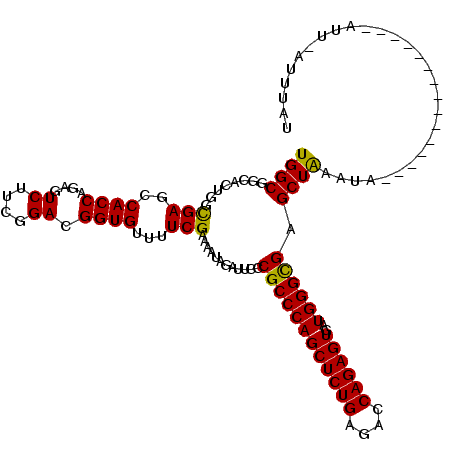
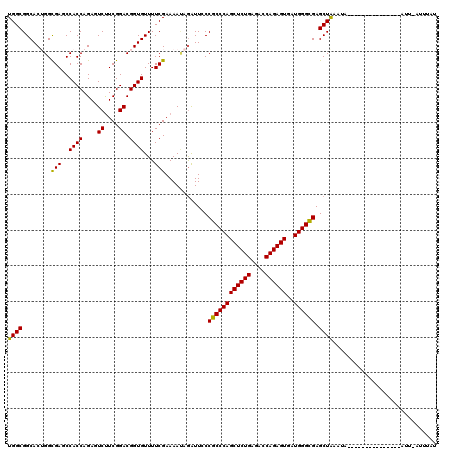
| Location | 1,119,395 – 1,119,500 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.04 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -28.13 |
| Energy contribution | -27.93 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1119395 105 - 27905053 AUGUACGAAC---------------CAUUUAGCUCGCCCAUUACUCUGGUCUCAGAGCUGGGCGGGAAUCUCUUUUCGAAAACACCGUCCGAAGACUCUGGUGGCUCACCUGUGCCGCCA .....((((.---------------.......((((((((...(((((....))))).))))))))........))))........(((....)))...((((((.(....).)))))). ( -37.29) >DroVir_CAF1 46977 105 - 1 AUAAAUGAAU---------------UACUUAGCUCGCCCAUCACUCUGGUAUCAGAGCUGGGCGGCAACUUGUUUUCGAAAACACCGUCCGAAGAAUCUGGUGGGUCGCCAGUGCCGCCA ..........---------------....((((((..(((......))).....))))))(((((((.....((((((...........))))))..((((((...))))))))))))). ( -34.10) >DroPse_CAF1 59612 104 - 1 ACACAA-AAU---------------UCUAUAGCUCGCCCAUUACUCUGGUCUCAGAGCUGGGCGGGAAUCUGUUUUCGAAAACACCGUCCGAAGACUCUGGUGGCUCGCCAGUGCCGCCA ......-...---------------.......((((((((...(((((....))))).)))))))).....(((((((...........)))))))...((((((.(....).)))))). ( -38.70) >DroGri_CAF1 60620 120 - 1 AAAAAAAAAAAUGUUUAUAUGCAAUUACUUAGCUCACCCAUCACUCUGGUCUCAGAGCUGGGCGGCAAUCUAUUUUCGAAAACACCGUCCGAAGAAUCUGGUGGGUCGCCAGUGCCGCCA ...........(((......)))......((((((..(((......))).....))))))(((((((.....((((((...........))))))..((((((...))))))))))))). ( -33.70) >DroMoj_CAF1 48067 108 - 1 AAAAAUCAAAAU---------C---AAAUCAGCUCGCCCAUCACUCUGGUCUCAGAGCUGGGCGGCAAUCUAUUUUCGAAAACACCGUCCGAAGAAUCUGGUGGAUCACCAGUGCCGCCA ............---------.---....((((((..(((......))).....))))))(((((((.....((((((...........))))))..((((((...))))))))))))). ( -36.60) >DroAna_CAF1 50775 92 - 1 ----------------------------UUAGCUCGCCCAUCACUCUGGUGUCAGAGCUGGGCGGGAAUCUAUUUUCGAAAACACCGUCCGAGGACUCUGGUGGCUCACCUGUGCCGCCA ----------------------------....((((((((...(((((....))))).))))))))....................(((....)))...((((((.(....).)))))). ( -35.80) >consensus AAAAAA_AAA_______________UACUUAGCUCGCCCAUCACUCUGGUCUCAGAGCUGGGCGGCAAUCUAUUUUCGAAAACACCGUCCGAAGAAUCUGGUGGCUCACCAGUGCCGCCA .............................((((((..(((......))).....))))))(((((.......((((((...........))))))..((((((...))))))..))))). (-28.13 = -27.93 + -0.19)

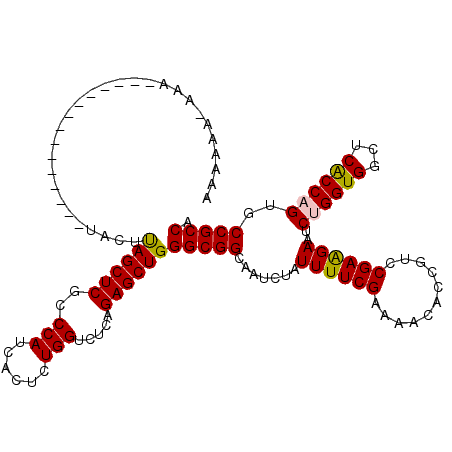
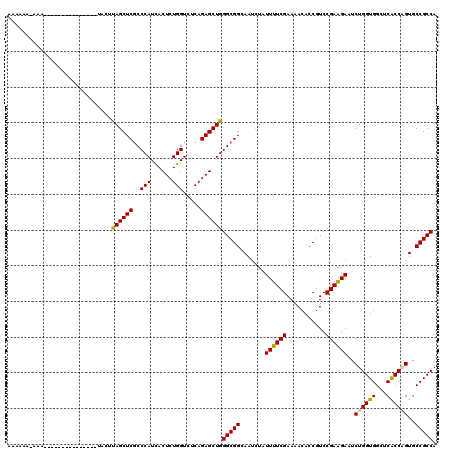
| Location | 1,119,435 – 1,119,529 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1119435 94 + 27905053 UUCGAAAAGAGAUUCCCGCCCAGCUCUGAGACCAGAGUAAUGGGCGAGCUAAAUG---GUUCGUACAUGAGUGUGGAAAGUGUAUAUAGGCAAGUUA ............((((((((((((((((....))))))..)))))(((((....)---))))..........).))))..(((......)))..... ( -26.00) >DroSec_CAF1 53122 94 + 1 UUCGAAAAGAGAUUCCCGCCCAGCUCUGAGACCAGAGUGAUGGGCGAGCUAUAUG---GUUCGUACAUGAGUGUGGAAAGUAUAUAUAGGCAAGAUA ................((((((((((((....))))))..))))))..(((((((---.(((.((((....)))))))....)))))))........ ( -28.00) >DroSim_CAF1 53663 94 + 1 UUCGAAAAGAGAUUCCCGCCCAGCUCUGAGACCAGAGUGAUGGGCGAGCUAUAUG---GUUCGUACAUGAGUGUGGAAAGUAUAUAUAGGCAAGAUA ................((((((((((((....))))))..))))))..(((((((---.(((.((((....)))))))....)))))))........ ( -28.00) >DroEre_CAF1 53703 89 + 1 UUCGAAAAUAGAUUCCCGCCCAGCUCUGAGACCAGAGUGAUGGGCGAGCUAUAUG---GU----ACAUAGGUAUGUAAAGUAUAUGUAGGCGGG-UA ..............((((((((((((((....))))))..)))))...(((((((---.(----((((....))))).....)))))))..)))-.. ( -29.90) >DroYak_CAF1 74191 90 + 1 UUCGAAAAAAGAUUCCCGCCCAGCUCUGAAACCAGAGUAAUGGGCGAGCUAAAUG---GU----ACGUAAGUGUUUUAAGUAUAUGUAGGCGGGAUA .............(((((((((((((((....))))))..)))))..(((.....---((----((.((((...)))).)))).....))))))).. ( -29.10) >DroMoj_CAF1 48107 91 + 1 UUCGAAAAUAGAUUGCCGCCCAGCUCUGAGACCAGAGUGAUGGGCGAGCUGAUUUGAUUUUGAUUUUUUUUUAUAGA---GAUAUAUAU---GUAUA .((((((.((((((((((((((((((((....))))))..)))))).)).)))))).))))))..............---.........---..... ( -29.10) >consensus UUCGAAAAGAGAUUCCCGCCCAGCUCUGAGACCAGAGUGAUGGGCGAGCUAAAUG___GUUCGUACAUGAGUGUGGAAAGUAUAUAUAGGCAAGAUA ................((((((((((((....))))))..))))))................................................... (-19.20 = -19.20 + -0.00)

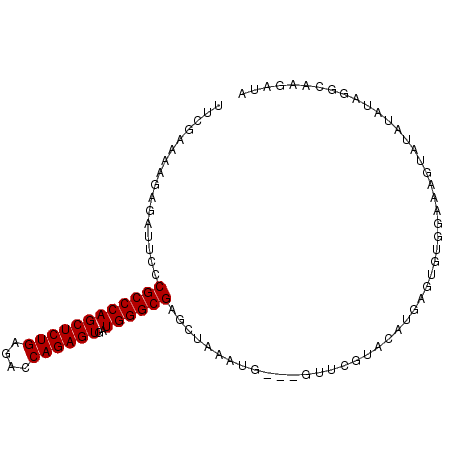
| Location | 1,119,435 – 1,119,529 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -23.53 |
| Consensus MFE | -19.48 |
| Energy contribution | -19.65 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1119435 94 - 27905053 UAACUUGCCUAUAUACACUUUCCACACUCAUGUACGAAC---CAUUUAGCUCGCCCAUUACUCUGGUCUCAGAGCUGGGCGGGAAUCUCUUUUCGAA ..................................((((.---.......((((((((...(((((....))))).))))))))........)))).. ( -23.69) >DroSec_CAF1 53122 94 - 1 UAUCUUGCCUAUAUAUACUUUCCACACUCAUGUACGAAC---CAUAUAGCUCGCCCAUCACUCUGGUCUCAGAGCUGGGCGGGAAUCUCUUUUCGAA ..................................((((.---.......((((((((...(((((....))))).))))))))........)))).. ( -23.69) >DroSim_CAF1 53663 94 - 1 UAUCUUGCCUAUAUAUACUUUCCACACUCAUGUACGAAC---CAUAUAGCUCGCCCAUCACUCUGGUCUCAGAGCUGGGCGGGAAUCUCUUUUCGAA ..................................((((.---.......((((((((...(((((....))))).))))))))........)))).. ( -23.69) >DroEre_CAF1 53703 89 - 1 UA-CCCGCCUACAUAUACUUUACAUACCUAUGU----AC---CAUAUAGCUCGCCCAUCACUCUGGUCUCAGAGCUGGGCGGGAAUCUAUUUUCGAA ..-((((((((..((((...(((((....))))----).---..))))((((..(((......))).....)))))))))))).............. ( -25.40) >DroYak_CAF1 74191 90 - 1 UAUCCCGCCUACAUAUACUUAAAACACUUACGU----AC---CAUUUAGCUCGCCCAUUACUCUGGUUUCAGAGCUGGGCGGGAAUCUUUUUUCGAA ..((((((((.....(((.(((.....))).))----).---.....(((((..(((......))).....)))))))))))))............. ( -24.50) >DroMoj_CAF1 48107 91 - 1 UAUAC---AUAUAUAUC---UCUAUAAAAAAAAAUCAAAAUCAAAUCAGCUCGCCCAUCACUCUGGUCUCAGAGCUGGGCGGCAAUCUAUUUUCGAA .....---.........---............................((.((((((...(((((....))))).)))))))).............. ( -20.20) >consensus UAUCCUGCCUAUAUAUACUUUCCACACUCAUGUACGAAC___CAUAUAGCUCGCCCAUCACUCUGGUCUCAGAGCUGGGCGGGAAUCUCUUUUCGAA .................................................((((((((...(((((....))))).)))))))).............. (-19.48 = -19.65 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:40 2006