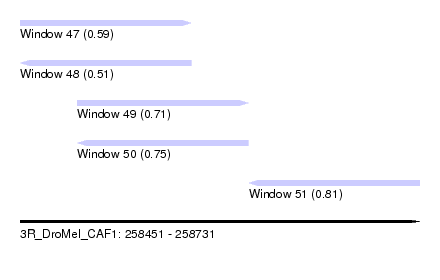
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 258,451 – 258,731 |
| Length | 280 |
| Max. P | 0.813606 |

| Location | 258,451 – 258,571 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.50 |
| Mean single sequence MFE | -43.13 |
| Consensus MFE | -30.51 |
| Energy contribution | -30.27 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
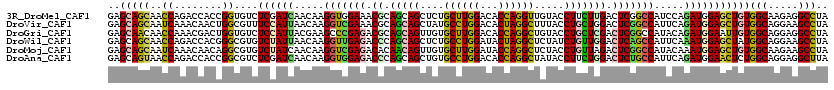
>3R_DroMel_CAF1 258451 120 + 27905053 CGUCAGCUGGAAGAGCUUCAGCAGCGCUGCAGCGUGGAACAACUGGUUGCCGGCCUUGCUAAAGACCAUGGCUAAUUUUUGGUAGUAGUUGGCCAUGGUUUUGGGCACGGGUGUCUUCUU ((.((((..(....(((.((((...)))).)))(.....)..)..)))).))((((((((.((((((((((((((((.........)))))))))))))))).)))).))))........ ( -48.10) >DroVir_CAF1 1938 120 + 1 CGUCAACUGAAACAGUUUAAGCAAAGCGGCUGCAUGGAAUAGCUGGUUACCAGCCUUGCUAAAUACCAUAGCCAGCUUCUGAUAGUAAUUGGCCAUGGUUUUGGGCACCGGCGUCUUCUU (((((((((...)))))..........((.(((........(((((...)))))....(((((.(((((.(((((((......)))...)))).)))))))))))))))))))....... ( -34.10) >DroGri_CAF1 1875 120 + 1 CGUCAACUGGAACAGUUUGAGCAAAGCGGCCGCAUGGAAGAGCUGAUUGCCGGCCUUGCUGAAAACCAUGGCAAGUUUCUGAUAAUAAUUUGCCAUGGUUUUGGGCACCGGCGUCUUCUU ........((((((((((...(...((....))...)..))))))...(((((...((((.((((((((((((((((.........)))))))))))))))).)))))))))...)))). ( -47.90) >DroWil_CAF1 2455 120 + 1 UGUUAGUUGGAACAACUUGAGCAAAGCAGCUGCAUGGAACAGUUGGUUGCCAGCCUUGCUGAAGACCAUUGCAAGUUUUUGGUAAUAGUUUGCCAUGGUCUUGGGCACGGGCGUCUUCUU ....(((((...)))))((.((((..((((((.......)))))).))))))((((((((.((((((((.((((..(.........)..)))).)))))))).)))).))))........ ( -42.30) >DroMoj_CAF1 2513 120 + 1 AGUCAACUGGAACAGCUUGAGCAAAGCGGCUGCAUGGAAGAGCUGAUUGCCAGCCUUGCUGAAGACCAUAGCCAAUUUCUGAUAAUAAUUAGCCAUGGUUUUUGGCACCGGCGUCUUCUU ........((((..((((.....))))(((.((.((((((.((((.....)))))))((..((((((((.((.((((.........)))).)).))))))))..)).)))))))))))). ( -37.50) >DroAna_CAF1 2509 120 + 1 AGUCAGCUGGAAGAGCUUCAGAAGAGCGGCAGCGUGGAACAACUGGUUGCCAGCCUUGCUGAAGACCAUCGCCAGUUUCUGAUAGUAAUUGGCCAUGGUUUUGGGCACCGGCGUCUUCUU ........((((((((((.....))))((((((..(......)..)))))).(((.((((.((((((((.(((((((.(.....).))))))).)))))))).))))..))).)))))). ( -48.90) >consensus CGUCAACUGGAACAGCUUGAGCAAAGCGGCUGCAUGGAACAGCUGGUUGCCAGCCUUGCUGAAGACCAUAGCCAGUUUCUGAUAAUAAUUGGCCAUGGUUUUGGGCACCGGCGUCUUCUU .......(((..(((((....(...((....))...)...)))))....)))(((.((((.((((((((.(((((((.........))))))).)))))))).))))..)))........ (-30.51 = -30.27 + -0.25)

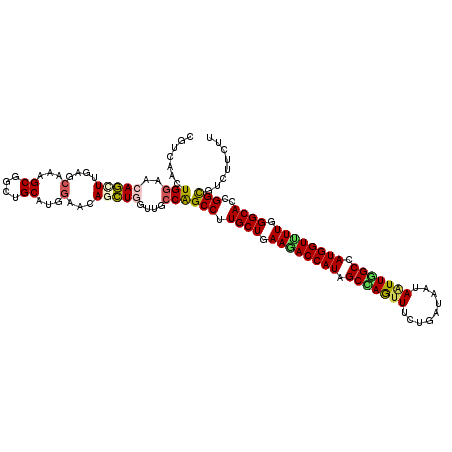
| Location | 258,451 – 258,571 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.50 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -26.77 |
| Energy contribution | -26.56 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 258451 120 - 27905053 AAGAAGACACCCGUGCCCAAAACCAUGGCCAACUACUACCAAAAAUUAGCCAUGGUCUUUAGCAAGGCCGGCAACCAGUUGUUCCACGCUGCAGCGCUGCUGAAGCUCUUCCAGCUGACG ..((((((..((.(((..((.((((((((...................)))))))).))..))).)).(((((....(((((........)))))..)))))..).)))))......... ( -32.21) >DroVir_CAF1 1938 120 - 1 AAGAAGACGCCGGUGCCCAAAACCAUGGCCAAUUACUAUCAGAAGCUGGCUAUGGUAUUUAGCAAGGCUGGUAACCAGCUAUUCCAUGCAGCCGCUUUGCUUAAACUGUUUCAGUUGACG (((.(((.((.(.(((.....((((((((((.((.(.....).)).)))))))))).........((((((...)))))).......))).).))))).))).(((((...))))).... ( -32.90) >DroGri_CAF1 1875 120 - 1 AAGAAGACGCCGGUGCCCAAAACCAUGGCAAAUUAUUAUCAGAAACUUGCCAUGGUUUUCAGCAAGGCCGGCAAUCAGCUCUUCCAUGCGGCCGCUUUGCUCAAACUGUUCCAGUUGACG ..((((..(((..(((..(((((((((((((...............)))))))))))))..))).)))((((.....((........)).))))))))..((((.(((...))))))).. ( -38.66) >DroWil_CAF1 2455 120 - 1 AAGAAGACGCCCGUGCCCAAGACCAUGGCAAACUAUUACCAAAAACUUGCAAUGGUCUUCAGCAAGGCUGGCAACCAACUGUUCCAUGCAGCUGCUUUGCUCAAGUUGUUCCAACUAACA ........(((..(((..((((((((.((((...............)))).))))))))..))).)))((((((.((.((((.....)))).))..)))).))(((((...))))).... ( -32.76) >DroMoj_CAF1 2513 120 - 1 AAGAAGACGCCGGUGCCAAAAACCAUGGCUAAUUAUUAUCAGAAAUUGGCUAUGGUCUUCAGCAAGGCUGGCAAUCAGCUCUUCCAUGCAGCCGCUUUGCUCAAGCUGUUCCAGUUGACU ..((((((...(((.......)))((((((((((.........)))))))))).))))))((((((((.(((.....((........)).)))))))))))..(((((...))))).... ( -37.90) >DroAna_CAF1 2509 120 - 1 AAGAAGACGCCGGUGCCCAAAACCAUGGCCAAUUACUAUCAGAAACUGGCGAUGGUCUUCAGCAAGGCUGGCAACCAGUUGUUCCACGCUGCCGCUCUUCUGAAGCUCUUCCAGCUGACU .((((((.((.(((((..((.(((((.((((.((.(.....).)).)))).))))).))..))..((((((.(((.....)))))).))))))))))))))..((((.....)))).... ( -36.60) >consensus AAGAAGACGCCGGUGCCCAAAACCAUGGCCAAUUACUAUCAGAAACUGGCCAUGGUCUUCAGCAAGGCUGGCAACCAGCUGUUCCAUGCAGCCGCUUUGCUCAAGCUGUUCCAGCUGACG ..((((((((....)).......((((((((...............))))))))))))))((((((((.(((.....((........)).)))))))))))..((((.....)))).... (-26.77 = -26.56 + -0.21)
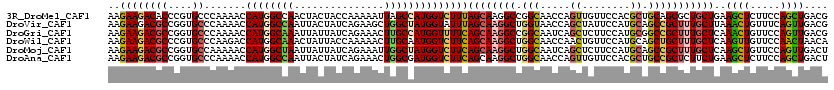
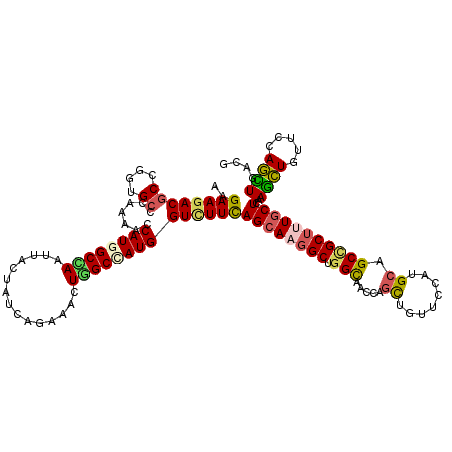
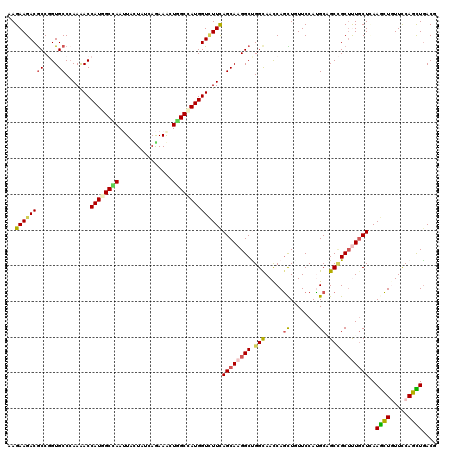
| Location | 258,491 – 258,611 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.17 |
| Mean single sequence MFE | -44.48 |
| Consensus MFE | -32.04 |
| Energy contribution | -30.80 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 258491 120 + 27905053 AACUGGUUGCCGGCCUUGCUAAAGACCAUGGCUAAUUUUUGGUAGUAGUUGGCCAUGGUUUUGGGCACGGGUGUCUUCUUGGACAGAGCCAUCAGGCCAUGGAUAUCCUCGAUUGCCUUG ...(((((.(((((((.....((((((((((((((((.........)))))))))))))))))))).))).(((((....))))).)))))..((((.((.((.....)).)).)))).. ( -47.20) >DroVir_CAF1 1978 120 + 1 AGCUGGUUACCAGCCUUGCUAAAUACCAUAGCCAGCUUCUGAUAGUAAUUGGCCAUGGUUUUGGGCACCGGCGUCUUCUUGGACAUGGCCAUCAGGCCGUGAAUGUCCUCAAUUGCCUUG .(((((...)))))..((((....(((((.(((((((......)))...)))).)))))....))))..((((.......((((((((((....))))....)))))).....))))... ( -43.80) >DroGri_CAF1 1915 120 + 1 AGCUGAUUGCCGGCCUUGCUGAAAACCAUGGCAAGUUUCUGAUAAUAAUUUGCCAUGGUUUUGGGCACCGGCGUCUUCUUGGACAUGGCCAUUAGGCCGUGAAUGUCCUCAAUGGCCUUG ........(((((...((((.((((((((((((((((.........)))))))))))))))).)))))))))((((....))))..((((((((((.((....)).))).)))))))... ( -56.70) >DroWil_CAF1 2495 120 + 1 AGUUGGUUGCCAGCCUUGCUGAAGACCAUUGCAAGUUUUUGGUAAUAGUUUGCCAUGGUCUUGGGCACGGGCGUCUUCUUGGAGAGAGCCAUCAGACCGUGAAUAUCCUCGAUGGCCUUG ........((((((((((((.((((((((.((((..(.........)..)))).)))))))).)))).))))((((...(((......)))..))))...............)))).... ( -41.10) >DroYak_CAF1 2542 120 + 1 AACUGGUUGCCAGCCUUGCUAAAGACCAUGGCUAAUUUUUGGUAAUAGUUGGCCAUUGUUUUGGGUACGGGUGUCUUUUUGGAUAAAGCCAUCAAGCCGUGGAUAUCCUCGAUUGCCCUG ..((((...))))...((((.(((((.((((((((((.........)))))))))).))))).))))((((.((..((..(((((...((((......)))).)))))..))..)))))) ( -36.30) >DroMoj_CAF1 2553 120 + 1 AGCUGAUUGCCAGCCUUGCUGAAGACCAUAGCCAAUUUCUGAUAAUAAUUAGCCAUGGUUUUUGGCACCGGCGUCUUCUUGGACAUGGCCAUCAGACCGUGAAUGUCCUCGAUUGCCUUG ..(((((.(((((((.(((..((((((((.((.((((.........)))).)).))))))))..)))..)))((((....)))).)))).)))))...(..(.((....)).)..).... ( -41.80) >consensus AGCUGGUUGCCAGCCUUGCUAAAGACCAUGGCAAAUUUCUGAUAAUAAUUGGCCAUGGUUUUGGGCACCGGCGUCUUCUUGGACAUAGCCAUCAGGCCGUGAAUAUCCUCGAUUGCCUUG .((((.....))))..((((.((((((((.(((((((.........))))))).)))))))).))))..(((((((...(((......)))..))))..(((......)))...)))... (-32.04 = -30.80 + -1.24)

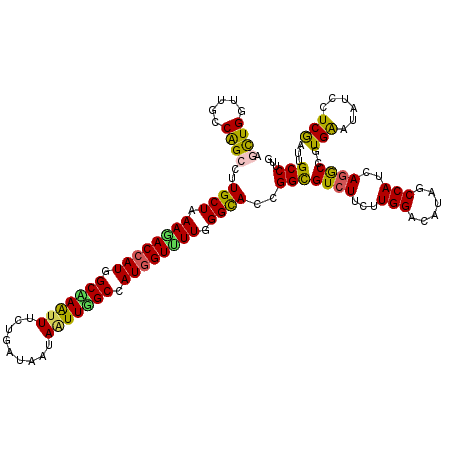
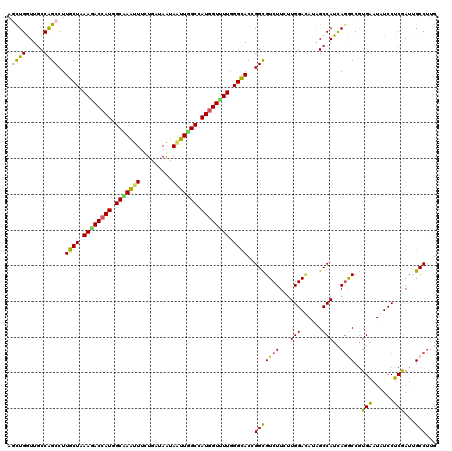
| Location | 258,491 – 258,611 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.17 |
| Mean single sequence MFE | -38.01 |
| Consensus MFE | -29.08 |
| Energy contribution | -28.98 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 258491 120 - 27905053 CAAGGCAAUCGAGGAUAUCCAUGGCCUGAUGGCUCUGUCCAAGAAGACACCCGUGCCCAAAACCAUGGCCAACUACUACCAAAAAUUAGCCAUGGUCUUUAGCAAGGCCGGCAACCAGUU ....((......((....)).((((((.((((...((((......)))).))))((..((.((((((((...................)))))))).))..)).))))))))........ ( -33.21) >DroVir_CAF1 1978 120 - 1 CAAGGCAAUUGAGGACAUUCACGGCCUGAUGGCCAUGUCCAAGAAGACGCCGGUGCCCAAAACCAUGGCCAAUUACUAUCAGAAGCUGGCUAUGGUAUUUAGCAAGGCUGGUAACCAGCU ...(((......((((((....((((....))))))))))..(....))))..(((..((.((((((((((.((.(.....).)).)))))))))).))..))).((((((...)))))) ( -43.60) >DroGri_CAF1 1915 120 - 1 CAAGGCCAUUGAGGACAUUCACGGCCUAAUGGCCAUGUCCAAGAAGACGCCGGUGCCCAAAACCAUGGCAAAUUAUUAUCAGAAACUUGCCAUGGUUUUCAGCAAGGCCGGCAAUCAGCU ...(((...(((((((((....((((....))))))))))........((((((((..(((((((((((((...............)))))))))))))..))...))))))..)))))) ( -48.86) >DroWil_CAF1 2495 120 - 1 CAAGGCCAUCGAGGAUAUUCACGGUCUGAUGGCUCUCUCCAAGAAGACGCCCGUGCCCAAGACCAUGGCAAACUAUUACCAAAAACUUGCAAUGGUCUUCAGCAAGGCUGGCAACCAACU ....((((...............((((..(((......)))...))))(((..(((..((((((((.((((...............)))).))))))))..))).)))))))........ ( -34.76) >DroYak_CAF1 2542 120 - 1 CAGGGCAAUCGAGGAUAUCCACGGCUUGAUGGCUUUAUCCAAAAAGACACCCGUACCCAAAACAAUGGCCAACUAUUACCAAAAAUUAGCCAUGGUCUUUAGCAAGGCUGGCAACCAGUU ....((......(((((.....((((....)))).)))))..((((((....((.......)).(((((...................))))).)))))).))..((((((...)))))) ( -24.21) >DroMoj_CAF1 2553 120 - 1 CAAGGCAAUCGAGGACAUUCACGGUCUGAUGGCCAUGUCCAAGAAGACGCCGGUGCCAAAAACCAUGGCUAAUUAUUAUCAGAAAUUGGCUAUGGUCUUCAGCAAGGCUGGCAAUCAGCU ...(((..((..((((((....((((....))))))))))..))....((((((((..((.(((((((((((((.........))))))))))))).))..))...)))))).....))) ( -43.40) >consensus CAAGGCAAUCGAGGACAUUCACGGCCUGAUGGCCAUGUCCAAGAAGACGCCCGUGCCCAAAACCAUGGCCAACUAUUACCAAAAACUAGCCAUGGUCUUCAGCAAGGCUGGCAACCAGCU ....((......(((((.....((((....)))).)))))........(((..(((..((.((((((((((...............)))))))))).))..))).)))..))........ (-29.08 = -28.98 + -0.11)

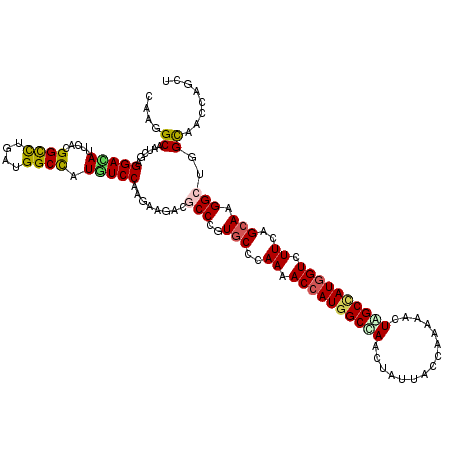
| Location | 258,611 – 258,731 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.61 |
| Mean single sequence MFE | -42.25 |
| Consensus MFE | -34.33 |
| Energy contribution | -33.67 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 258611 120 - 27905053 GAGCAGCAACCAGACCACCGGUGUCUCGAUCAACAAGGUGGAAACGCAGCAGCUCUGCUUGGACACCAGGUUGUACCUUCUUGACUCGGCCAUCCAGAUGGAGCUGUGGCAAGAGGCCUA .....((((((.(.....)(((((((...........(((....)))((((....)))).))))))).))))))..(((((((.(.((((..(((....))))))).).))))))).... ( -42.90) >DroVir_CAF1 2098 120 - 1 GAGCAGCAAUCAAACAACUGGCGUUUCCAUUAACAAGGUCGAAACGCAGCAGCUAUGCCUGGACACUAGGCUUUACCUGCUGGACUCGGCCAUUCAGAUGGAGCUGUGGCAGGAAGCCUA ..(((((..(((......)))....((((((.....((((((..(.((((((.((.((((((...))))))..)).)))))))..)))))).....)))))))))))(((.....))).. ( -44.00) >DroGri_CAF1 2035 120 - 1 GAGCAACAACCAAACGACUGGUGUCUCCAUUACGAAGCCCGAGACGCAGCAGUUGUGCUUGGACACCAGGCUGUACCUGCUCGACUCGGCCAUACAGAUGGAAUUGUGGCAGGAGGCCUA ........((((......))))((((((.((((((...(((((.((.(((((....((((((...)))))).....))))))).)))))((((....))))..))))))..))))))... ( -42.50) >DroWil_CAF1 2615 120 - 1 GAGCAGCAACCAGACCACGGGCGUGUCUAUUAACAAGGUUGAGACCCAGCAGCUCUGCCUGGAUACUAGGCUCUAUCUGUUGGACUCAGCCAUUCAAAUGGAGCUAUGGCAGGAAGCCUA .....((..((.(.(((..(((...((((((.....(((((((..(((((((....((((((...)))))).....))))))).))))))).....))))))))).)))).))..))... ( -41.60) >DroMoj_CAF1 2673 120 - 1 GAGCAGCAAUCAAACAACAGGCGUGUCUAUCAACAAGGUCGAGACACAACAGUUGUGCUUGGAUACCAGGCUCUACCUGUUAGACUCGGCCAUACAAAUGGAGCUGUGGCAAGAAGCCUA ..(((((........((((((.(((((((((.....)))..)))))).......(.((((((...)))))).)..))))))........((((....)))).)))))(((.....))).. ( -37.10) >DroAna_CAF1 2669 120 - 1 GAGCAGUAACCAGACCACCGGCGUCUCGAUCAACAAGGUGGAGACCCAGCAGCUGUGCCUGGACACCAGGCUAUACCUUCUGGACUCUGCCAUUCAGAUGGAACUCUGGCAGGAGGCUUA ((((.....(((((.....((.(((((.(((.....))).)))))))...((.(((((((((...)))))).))).))))))).(((((((((((.....)))...))))).))))))). ( -45.40) >consensus GAGCAGCAACCAAACCACCGGCGUCUCCAUCAACAAGGUCGAGACGCAGCAGCUGUGCCUGGACACCAGGCUCUACCUGCUGGACUCGGCCAUUCAGAUGGAGCUGUGGCAGGAAGCCUA ..(((((..((........))....((((((.....(((((((.((.(((((....((((((...)))))).....))))))).))))))).....)))))))))))(((.....))).. (-34.33 = -33.67 + -0.66)
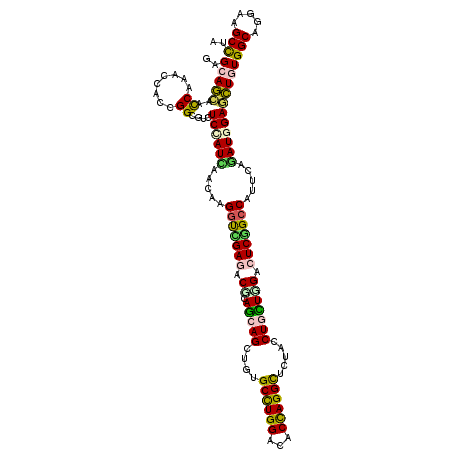
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:49 2006