
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,775,494 – 7,775,694 |
| Length | 200 |
| Max. P | 0.996120 |

| Location | 7,775,494 – 7,775,614 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -36.26 |
| Consensus MFE | -31.52 |
| Energy contribution | -31.64 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7775494 120 - 27905053 UGUGCCCAUGGCCAUGUUGUCCAAGUCCAUGGACGUGGGCAUGCUUGUUGUAGCCCAAAUCGGCGUUUCCGUUUCCACCAGGAAACAUCUCUGCUUGUAGUUCGAAUAUGCUCUUUAAAU .(((((((((.(((((...........))))).)))))))))((.((((...(((......)))......((((((....))))))..................)))).))......... ( -35.90) >DroSec_CAF1 72724 120 - 1 UGUGCCCAUGGCCAUGUUGUCCAUGUCCGUGGACGUGGGCGUGCUUGUUGUAGCCCAAAUCGGCGUUUCCAUUUCCACCAGGAAACAUCUCUGCUUGUAGUUCGAAUAUGCUCUUUAAAU ...((((((((.((((.....)))).)))))((..(((((.(((.....))))))))..)))))((((((..........))))))......((.(((.......))).))......... ( -35.30) >DroSim_CAF1 75967 120 - 1 UGUGCCCAUGGCCAUGUUGUCCGUGCCCGUGGACGUGGGCGUGCUUGUUGUAGCCCAAAUCGGCGUUUCCAUUUCCACCAGGAAACAUCUCUGCUUGUAGUUCGAAUAUGCUCUUUAAAU ...((((((((.((((.....)))).)))))((..(((((.(((.....))))))))..)))))((((((..........))))))......((.(((.......))).))......... ( -34.90) >DroEre_CAF1 73172 119 - 1 UGUGCCCAUGGCCAUGUUGCCCGUGUCCAUGGACAUGGGCGUGCUUGUUGUAGCCCAAAUCGGCGUUUCCAUUUCCACCAGGAAACAUCUCUGCUU-UAGCUUGAAUAUGUUUUUUGAAU .(((...((((..((((((((((((((....)))))))))(.(((......))).).....)))))..))))...)))((((((((((.((.((..-..))..))..))))))))))... ( -39.70) >DroYak_CAF1 73552 120 - 1 UGUGGCCAUGGCCAUGUUGUCCGUGUCCAUGGACGUGGGCAUGCUUGUUGUAACCCAAAUCGGCGUUUCCGUUUCCACCGGGAAACAUCUCUGCUUGUAUCUUGAUUAUGCUUUUAGAAU .(((((....)))))...((((((....))))))..(((.(((((.(((........))).))))).)))((((((....))))))...((((...((((.......))))...)))).. ( -35.50) >consensus UGUGCCCAUGGCCAUGUUGUCCGUGUCCAUGGACGUGGGCGUGCUUGUUGUAGCCCAAAUCGGCGUUUCCAUUUCCACCAGGAAACAUCUCUGCUUGUAGUUCGAAUAUGCUCUUUAAAU ...((((((((.((((.....)))).)))))((..(((((.(((.....))))))))..)))))((((((..........)))))).................................. (-31.52 = -31.64 + 0.12)



| Location | 7,775,534 – 7,775,654 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -39.94 |
| Consensus MFE | -36.64 |
| Energy contribution | -36.80 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7775534 120 + 27905053 UGGUGGAAACGGAAACGCCGAUUUGGGCUACAACAAGCAUGCCCACGUCCAUGGACUUGGACAACAUGGCCAUGGGCACAACCAUAAUCACAAUCAGUUCCUGGCGCAGCCCCCACCACC ((((((....((...((((((((((((((......))).((((((.(.(((((...........))))).).))))))...))).))))((.....))....))))...)).)))))).. ( -42.10) >DroSec_CAF1 72764 120 + 1 UGGUGGAAAUGGAAACGCCGAUUUGGGCUACAACAAGCACGCCCACGUCCACGGACAUGGACAACAUGGCCAUGGGCACAACCAUAAUCACACUCAGUUCCUGGCGCAGCCCCCGCCGCC .(((((....((...(((((...(((((............))))).(((((.((.((((.....)))).)).)))))........................)))))...))....))))) ( -40.30) >DroSim_CAF1 76007 120 + 1 UGGUGGAAAUGGAAACGCCGAUUUGGGCUACAACAAGCACGCCCACGUCCACGGGCACGGACAACAUGGCCAUGGGCACAACCAUAAUCACACUCAGUUCCUGGCGCAGCCCCCGCCGCC .(((((....((...((((((((((((((......)))..(((((((....))(((.((.......))))).)))))....))).))))((.....))....))))...))....))))) ( -38.90) >DroEre_CAF1 73211 120 + 1 UGGUGGAAAUGGAAACGCCGAUUUGGGCUACAACAAGCACGCCCAUGUCCAUGGACACGGGCAACAUGGCCAUGGGCACAAUCAUAAUCACAAUCAGUUCCUGGCACAGCCCCCGCCGCC .(((((....(....)........(((((...........((((.((((....)))).))))......((((.((((...................)))).))))..))))))))))... ( -42.41) >DroYak_CAF1 73592 120 + 1 CGGUGGAAACGGAAACGCCGAUUUGGGUUACAACAAGCAUGCCCACGUCCAUGGACACGGACAACAUGGCCAUGGCCACAACCAUAAUCACAAUCAGUUCCUGGCACAGCCCCCGCCGCC ((((((....((((..((((...(((((............))))).(((....))).))).)....((((....))))...................)))).(((...))).)))))).. ( -36.00) >consensus UGGUGGAAAUGGAAACGCCGAUUUGGGCUACAACAAGCACGCCCACGUCCAUGGACACGGACAACAUGGCCAUGGGCACAACCAUAAUCACAAUCAGUUCCUGGCGCAGCCCCCGCCGCC ((((((....(....)((((...(((((............))))).((((((((.((.(.....).)).))))))))........................)))).......)))))).. (-36.64 = -36.80 + 0.16)

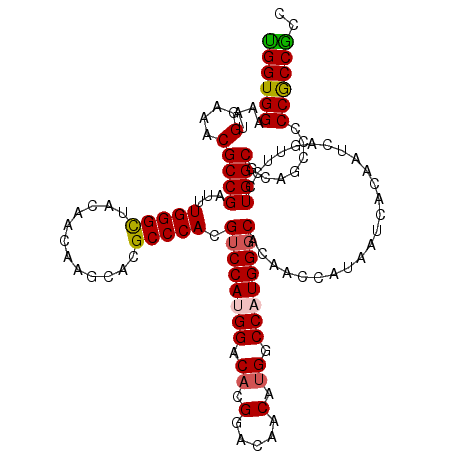

| Location | 7,775,534 – 7,775,654 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -48.84 |
| Consensus MFE | -43.02 |
| Energy contribution | -42.82 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7775534 120 - 27905053 GGUGGUGGGGGCUGCGCCAGGAACUGAUUGUGAUUAUGGUUGUGCCCAUGGCCAUGUUGUCCAAGUCCAUGGACGUGGGCAUGCUUGUUGUAGCCCAAAUCGGCGUUUCCGUUUCCACCA (((((..(.((..(((((.(((.....(((.(((((((((..(....)..))))))..)))))).)))...((..(((((.(((.....))))))))..)))))))..)).)..))))). ( -52.80) >DroSec_CAF1 72764 120 - 1 GGCGGCGGGGGCUGCGCCAGGAACUGAGUGUGAUUAUGGUUGUGCCCAUGGCCAUGUUGUCCAUGUCCGUGGACGUGGGCGUGCUUGUUGUAGCCCAAAUCGGCGUUUCCAUUUCCACCA ...((.((((((((((.((((.((..........((((((..(....)..))))))..(((((((((....))))))))))).)))).)))))))).....((.....))....)).)). ( -50.60) >DroSim_CAF1 76007 120 - 1 GGCGGCGGGGGCUGCGCCAGGAACUGAGUGUGAUUAUGGUUGUGCCCAUGGCCAUGUUGUCCGUGCCCGUGGACGUGGGCGUGCUUGUUGUAGCCCAAAUCGGCGUUUCCAUUUCCACCA .(.((((.((((.(((((((...))).))))...((((((..(....)..))))))..)))).)))))(((((.(((((..((((.(((........))).))))..))))).))))).. ( -47.40) >DroEre_CAF1 73211 120 - 1 GGCGGCGGGGGCUGUGCCAGGAACUGAUUGUGAUUAUGAUUGUGCCCAUGGCCAUGUUGCCCGUGUCCAUGGACAUGGGCGUGCUUGUUGUAGCCCAAAUCGGCGUUUCCAUUUCCACCA ...((.((((((((.((((((.((.(((..(....)..)))))..)).))))((.((((((((((((....)))))))))).)).))...)))))).....((.....))....)).)). ( -44.90) >DroYak_CAF1 73592 120 - 1 GGCGGCGGGGGCUGUGCCAGGAACUGAUUGUGAUUAUGGUUGUGGCCAUGGCCAUGUUGUCCGUGUCCAUGGACGUGGGCAUGCUUGUUGUAACCCAAAUCGGCGUUUCCGUUUCCACCG ((..((((..((...(((....((.....))((((..(((((..((((((.((((...((((((....)))))))))).))))...))..)))))..)))))))))..))))..)).... ( -48.50) >consensus GGCGGCGGGGGCUGCGCCAGGAACUGAUUGUGAUUAUGGUUGUGCCCAUGGCCAUGUUGUCCGUGUCCAUGGACGUGGGCGUGCUUGUUGUAGCCCAAAUCGGCGUUUCCAUUUCCACCA ...((.(((((((((..((((.............((((((..(....)..)))))).((((((((((....))))))))))..))))..))))))).....((.....))....)).)). (-43.02 = -42.82 + -0.20)



| Location | 7,775,574 – 7,775,694 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -43.43 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.12 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7775574 120 + 27905053 GCCCACGUCCAUGGACUUGGACAACAUGGCCAUGGGCACAACCAUAAUCACAAUCAGUUCCUGGCGCAGCCCCCACCACCCCCCACACAUUUUUCACUGCCCUCCGGGGGCGGUCAGGGC ((((..(((((......)))))....(((((((((......))))...........((((((((.((((...........................))))...))))))))))))))))) ( -41.33) >DroSec_CAF1 72804 120 + 1 GCCCACGUCCACGGACAUGGACAACAUGGCCAUGGGCACAACCAUAAUCACACUCAGUUCCUGGCGCAGCCCCCGCCGCCCCCCACACAUUUUCCAAUGCCCCCGGGCGGCGGUCAGGGC (((((.(((((.((.((((.....)))).)).)))))............((.....))...))).)).(((((((((((((......((((....)))).....)))))))))...)))) ( -46.10) >DroSim_CAF1 76047 120 + 1 GCCCACGUCCACGGGCACGGACAACAUGGCCAUGGGCACAACCAUAAUCACACUCAGUUCCUGGCGCAGCCCCCGCCGCCCCCCACACAUUUUCCAAUGCCCCCGGGCGGCGGUCAGGGC ((((........((((..(.....).((((((.((((...................)))).)))).))))))(((((((((......((((....)))).....)))))))))...)))) ( -44.41) >DroEre_CAF1 73251 120 + 1 GCCCAUGUCCAUGGACACGGGCAACAUGGCCAUGGGCACAAUCAUAAUCACAAUCAGUUCCUGGCACAGCCCCCGCCGCCACACACACAUUUUCCAAUGCCCCCGGGCGGGGGUCAGGGU (((((((.(((((...........))))).))))))).....................(((((.....((((((((((.....)...((((....))))......)))))))))))))). ( -44.70) >DroYak_CAF1 73632 120 + 1 GCCCACGUCCAUGGACACGGACAACAUGGCCAUGGCCACAACCAUAAUCACAAUCAGUUCCUGGCACAGCCCCCGCCGCCACCCCCUCAUUUUGCAAUGCCCCCGGGCGGGGGUCAGGGC ((((..(((((.((((..(.....).((((....))))..................)))).))).)).(((((((((............................)))))))))..)))) ( -40.59) >consensus GCCCACGUCCAUGGACACGGACAACAUGGCCAUGGGCACAACCAUAAUCACAAUCAGUUCCUGGCGCAGCCCCCGCCGCCCCCCACACAUUUUCCAAUGCCCCCGGGCGGCGGUCAGGGC ((((..((((........))))....(((((..((((....(((.....((.....))...)))....))))...(((((.......((((....))))......))))).))))))))) (-33.00 = -33.12 + 0.12)



| Location | 7,775,574 – 7,775,694 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -56.16 |
| Consensus MFE | -47.20 |
| Energy contribution | -48.80 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7775574 120 - 27905053 GCCCUGACCGCCCCCGGAGGGCAGUGAAAAAUGUGUGGGGGGUGGUGGGGGCUGCGCCAGGAACUGAUUGUGAUUAUGGUUGUGCCCAUGGCCAUGUUGUCCAAGUCCAUGGACGUGGGC (((((.(((((((((...(.(((.(....).))).).))))))))).))))).((((((...((.....)).....))).)))(((((((.(((((...........))))).))))))) ( -56.60) >DroSec_CAF1 72804 120 - 1 GCCCUGACCGCCGCCCGGGGGCAUUGGAAAAUGUGUGGGGGGCGGCGGGGGCUGCGCCAGGAACUGAGUGUGAUUAUGGUUGUGCCCAUGGCCAUGUUGUCCAUGUCCGUGGACGUGGGC ((((...(((((((((..(.(((((....))))).)...))))))))))))).(((((((...))).))))...((((((..(....)..))))))..(((((((((....))))))))) ( -61.40) >DroSim_CAF1 76047 120 - 1 GCCCUGACCGCCGCCCGGGGGCAUUGGAAAAUGUGUGGGGGGCGGCGGGGGCUGCGCCAGGAACUGAGUGUGAUUAUGGUUGUGCCCAUGGCCAUGUUGUCCGUGCCCGUGGACGUGGGC ((((((.(((((((((..(.(((((....))))).)...)))))))))((((.(((((((...))).))))(((((((((..(....)..))))))..)))...)))).....)).)))) ( -56.60) >DroEre_CAF1 73251 120 - 1 ACCCUGACCCCCGCCCGGGGGCAUUGGAAAAUGUGUGUGUGGCGGCGGGGGCUGUGCCAGGAACUGAUUGUGAUUAUGAUUGUGCCCAUGGCCAUGUUGCCCGUGUCCAUGGACAUGGGC .......((((((((((.(.(((((....))))).).)).))))).)))(((((((...((.((.(((..(....)..))))).))))))))).....(((((((((....))))))))) ( -54.70) >DroYak_CAF1 73632 120 - 1 GCCCUGACCCCCGCCCGGGGGCAUUGCAAAAUGAGGGGGUGGCGGCGGGGGCUGUGCCAGGAACUGAUUGUGAUUAUGGUUGUGGCCAUGGCCAUGUUGUCCGUGUCCAUGGACGUGGGC (((.(.(((((((((....)))(((....)))..)))))).).)))...((((((((((.((..((((....))))...)).))).))))))).....(((((((((....))))))))) ( -51.50) >consensus GCCCUGACCGCCGCCCGGGGGCAUUGGAAAAUGUGUGGGGGGCGGCGGGGGCUGCGCCAGGAACUGAUUGUGAUUAUGGUUGUGCCCAUGGCCAUGUUGUCCGUGUCCAUGGACGUGGGC .(((((.((((((((((...(((((....))))).)))))))))))))))((((.((((((.((.((((((....))))))))..)).)))))).)).(((((((((....))))))))) (-47.20 = -48.80 + 1.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:22 2006