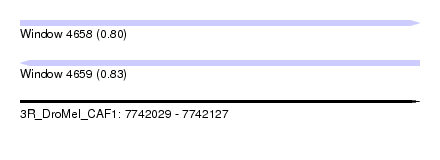
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,742,029 – 7,742,127 |
| Length | 98 |
| Max. P | 0.827723 |

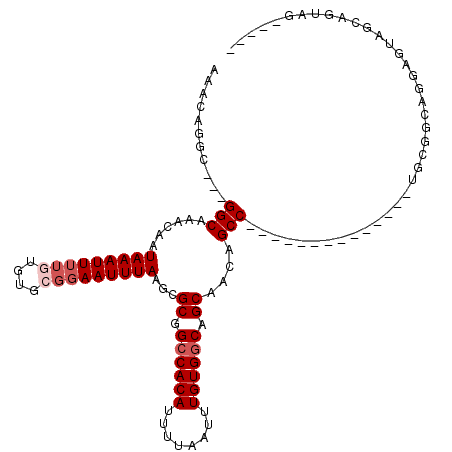
| Location | 7,742,029 – 7,742,127 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.60 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -17.64 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7742029 98 + 27905053 -----CUACUGCUACUCCUGCCGCA--------------GGCUGUUGCUGCCACAAAUUAAAAUGUGGCCGCGCUUAAAUUCCGCACACACAAUUUAUUGUUUGCCGCCGCCUGUUU -----.....((.......)).(((--------------(((.((.((.((((((........)))))).))...........(((.(((........))).))).)).)))))).. ( -27.30) >DroSec_CAF1 39134 98 + 1 -----CUACUGCUAUUCCUGCCGCA--------------GGCUGUUGCUGCCACAAAUUAAAAUGUGUCCGCGCUUAAAUUCCGCACACAAAAUUUAUUGUUUGCCGCCGCCUGUUU -----.....((.......)).(((--------------(((.((.((.(.((((........)))).).))...........(((.((((......)))).))).)).)))))).. ( -21.40) >DroSim_CAF1 41054 98 + 1 -----CUACUGCUACUCCUGCCGCA--------------GGCUGUUGCUGCCACAAAUUAAAAUGUGGCCGCGCUUAAAUUCCGCACACAAAAUUUAUUGUUUGCCGCCGCCUGUUU -----.....((.......)).(((--------------(((.((.((.((((((........)))))).))...........(((.((((......)))).))).)).)))))).. ( -28.50) >DroEre_CAF1 39148 109 + 1 -----CUCCUGUUACUCCUGCGGCAGCGCCAGCGGCGGCGGCUGUUGCUGCCACAAAUUAAAAUGUGGCUGCGCUUAAAUUCCCCACACAAAAUUUAUUGUUUGCC---GCCUGUUU -----..............(((((((((((((((((....)))))))..((((((........)))))).)))).............((((......)))).))))---))...... ( -35.90) >DroYak_CAF1 39414 111 + 1 UACUACUACCGUUACUCCUGCGGC---GACGGCGGCGGCGGCUGUUGCUGCCACAAAUUAAAAUGUGGCCGCGCUUAAAUUCCCCACACAAAAUUUAUUGUUUGCC---GCCUGUUU ........((((.......)))).---(((((((((((((((....))))))...........(((((...............)))))...............)))---))).))). ( -34.66) >DroAna_CAF1 52841 80 + 1 --------------CUCCUGC------------------GGCUGUUGCUGCCACAAAUUAAAAUGUGGCCGCGCUUAAAUUCUGC-CGC-AAAUUUAUUGUCUGCC---GCCAGCUU --------------...((((------------------(((....((.((((((........)))))).))((.........))-...-.............)))---).)))... ( -22.80) >consensus _____CUACUGCUACUCCUGCCGCA______________GGCUGUUGCUGCCACAAAUUAAAAUGUGGCCGCGCUUAAAUUCCGCACACAAAAUUUAUUGUUUGCC___GCCUGUUU .......................................(((....((.((((((........)))))).))...........(((.((((......)))).)))....)))..... (-17.64 = -18.50 + 0.86)

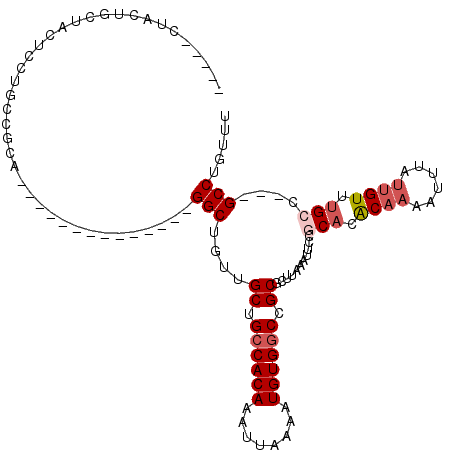
| Location | 7,742,029 – 7,742,127 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.60 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -19.97 |
| Energy contribution | -20.80 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7742029 98 - 27905053 AAACAGGCGGCGGCAAACAAUAAAUUGUGUGUGCGGAAUUUAAGCGCGGCCACAUUUUAAUUUGUGGCAGCAACAGCC--------------UGCGGCAGGAGUAGCAGUAG----- ...(..((.(((((......((((((.((....)).))))))...((.((((((........)))))).))....)))--------------.)).))..)...........----- ( -30.80) >DroSec_CAF1 39134 98 - 1 AAACAGGCGGCGGCAAACAAUAAAUUUUGUGUGCGGAAUUUAAGCGCGGACACAUUUUAAUUUGUGGCAGCAACAGCC--------------UGCGGCAGGAAUAGCAGUAG----- ...(..((.(((((......(((((((((....)))))))))...((...((((........))))...))....)))--------------.)).))..)...........----- ( -26.10) >DroSim_CAF1 41054 98 - 1 AAACAGGCGGCGGCAAACAAUAAAUUUUGUGUGCGGAAUUUAAGCGCGGCCACAUUUUAAUUUGUGGCAGCAACAGCC--------------UGCGGCAGGAGUAGCAGUAG----- ...(..((.(((((......(((((((((....)))))))))...((.((((((........)))))).))....)))--------------.)).))..)...........----- ( -32.20) >DroEre_CAF1 39148 109 - 1 AAACAGGC---GGCAAACAAUAAAUUUUGUGUGGGGAAUUUAAGCGCAGCCACAUUUUAAUUUGUGGCAGCAACAGCCGCCGCCGCUGGCGCUGCCGCAGGAGUAACAGGAG----- ...(..((---(((......((((((((......))))))))(((((.((((((........)))))).....((((.......))))))))))))))..)...........----- ( -38.10) >DroYak_CAF1 39414 111 - 1 AAACAGGC---GGCAAACAAUAAAUUUUGUGUGGGGAAUUUAAGCGCGGCCACAUUUUAAUUUGUGGCAGCAACAGCCGCCGCCGCCGUC---GCCGCAGGAGUAACGGUAGUAGUA .....(((---(((......((((((((......))))))))...((.((((((........)))))).))....))))))((..((((.---.((....).)..))))..)).... ( -40.80) >DroAna_CAF1 52841 80 - 1 AAGCUGGC---GGCAGACAAUAAAUUU-GCG-GCAGAAUUUAAGCGCGGCCACAUUUUAAUUUGUGGCAGCAACAGCC------------------GCAGGAG-------------- ...((.((---(((......(((((((-...-...)))))))...((.((((((........)))))).))....)))------------------)).))..-------------- ( -28.10) >consensus AAACAGGC___GGCAAACAAUAAAUUUUGUGUGCGGAAUUUAAGCGCGGCCACAUUUUAAUUUGUGGCAGCAACAGCC______________UGCGGCAGGAGUAGCAGUAG_____ ...........(((......(((((((((....)))))))))...((.((((((........)))))).))....)))....................................... (-19.97 = -20.80 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:06 2006