
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
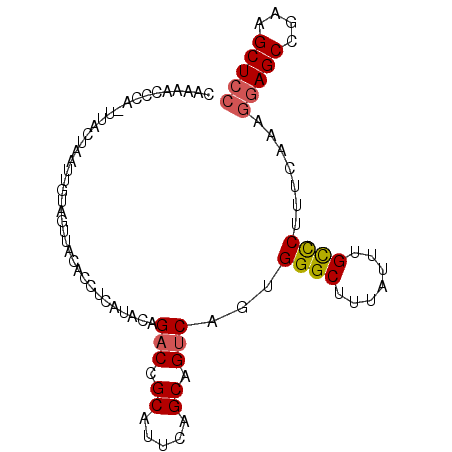
| Location | 7,704,752 – 7,704,884 |
| Length | 132 |
| Max. P | 0.980945 |

| Location | 7,704,752 – 7,704,844 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -16.34 |
| Energy contribution | -16.57 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7704752 92 + 27905053 CAAAGCCCA-UUACUAAUUGUAGUUACACCUCAUUUAGACCGCAUUCAGCAGUCAGUGGGCUUUGUUUGCCCUUUCAAAGGAGCCGAGGCUCC (((((((((-((((((....)))).............(((.((.....)).))))))))))))))..............(((((....))))) ( -30.80) >DroPse_CAF1 48609 93 + 1 CCGAAUUGAUUGACUAAUUCGAAUCCCCUCCCGCACAGACCGCCUUCAGCAGCCAGUGGGCGUUAUUCGUUCUUUCGCGGGAGCCACAGCUGC .(((((((......)))))))......(((((((..(((.((.....(((.(((....))))))...)).)))...))))))).......... ( -25.80) >DroSec_CAF1 1694 92 + 1 UAAAGCCCA-UUACUAAUUGUAGUUACACUUCAUACAGACCGCAUUUAGCAGUCAGUGGGCUUUGUUUGCCCUUUCAAAGGAGCCGACGCUCC (((((((((-((......((((...........))))(((.((.....)).))))))))))))))..............(((((....))))) ( -27.70) >DroSim_CAF1 1690 92 + 1 UAAAGCCCA-UUACUUAUUGUAGCUACACCUCAUACAGACCGCAUUUAGCAGUCAGUGGGCUUUGUUUGCCCUUUCAAAGGAGCCGACGCUCC (((((((((-((......((((...........))))(((.((.....)).))))))))))))))..............(((((....))))) ( -27.70) >DroEre_CAF1 1777 92 + 1 UGGAACCAA-UUACUAAUUGUAGUUAUGCUUCAUUUAGACCGCGUUCAGCAGUCAGUGGGCUCUAUUUGCCCUUUCGCAAGAGCCCCAGCUCC .(((...((-((((.....))))))..(((.......(((.((.....)).))).(.(((((((...(((......))))))))))))))))) ( -27.70) >DroYak_CAF1 1742 92 + 1 CGUAACCAA-UUACUAAUCGUAGUCAUACUUCGUUUAGACCGCGUUUAGCAGUCAGUGGGCAUUAUUUGCCCUUUCACAGGAGCCCCAGCUCC .((.((..(-((.((((.((..(((............)))..)).)))).)))..))(((((.....)))))....)).(((((....))))) ( -24.80) >consensus CAAAACCCA_UUACUAAUUGUAGUUACACCUCAUACAGACCGCAUUCAGCAGUCAGUGGGCUUUAUUUGCCCUUUCAAAGGAGCCGAAGCUCC .....................................(((.((.....)).)))...((((.......)))).......(((((....))))) (-16.34 = -16.57 + 0.22)


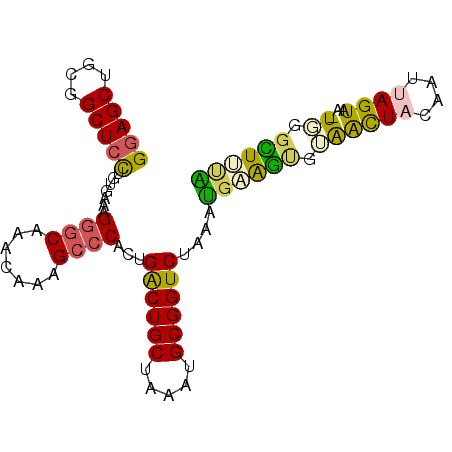
| Location | 7,704,752 – 7,704,844 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -23.28 |
| Energy contribution | -22.62 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7704752 92 - 27905053 GGAGCCUCGGCUCCUUUGAAAGGGCAAACAAAGCCCACUGACUGCUGAAUGCGGUCUAAAUGAGGUGUAACUACAAUUAGUAA-UGGGCUUUG (((((....)))))..............(((((((((..((((((.....))))))(((.((((......)).)).)))....-))))))))) ( -33.80) >DroPse_CAF1 48609 93 - 1 GCAGCUGUGGCUCCCGCGAAAGAACGAAUAACGCCCACUGGCUGCUGAAGGCGGUCUGUGCGGGAGGGGAUUCGAAUUAGUCAAUCAAUUCGG ..........(((((((....(..((.....))..)((.((((((.....)))))).)))))))))......((((((........)))))). ( -27.60) >DroSec_CAF1 1694 92 - 1 GGAGCGUCGGCUCCUUUGAAAGGGCAAACAAAGCCCACUGACUGCUAAAUGCGGUCUGUAUGAAGUGUAACUACAAUUAGUAA-UGGGCUUUA (((((....))))).......((((.......))))((.((((((.....)))))).)).((((((.((((((....))))..-)).)))))) ( -31.50) >DroSim_CAF1 1690 92 - 1 GGAGCGUCGGCUCCUUUGAAAGGGCAAACAAAGCCCACUGACUGCUAAAUGCGGUCUGUAUGAGGUGUAGCUACAAUAAGUAA-UGGGCUUUA (((((....))))).......((((.......))))((.((((((.....)))))).)).((((((.((..(((.....))).-)).)))))) ( -32.10) >DroEre_CAF1 1777 92 - 1 GGAGCUGGGGCUCUUGCGAAAGGGCAAAUAGAGCCCACUGACUGCUGAACGCGGUCUAAAUGAAGCAUAACUACAAUUAGUAA-UUGGUUCCA ((((((.((((((((((......)))...)))))))...((((((.....)))))).............((((....))))..-..)))))). ( -33.00) >DroYak_CAF1 1742 92 - 1 GGAGCUGGGGCUCCUGUGAAAGGGCAAAUAAUGCCCACUGACUGCUAAACGCGGUCUAAACGAAGUAUGACUACGAUUAGUAA-UUGGUUACG (((((....))))).((((..(((((.....))))).(..(.((((((.((..(((............)))..)).)))))).-)..))))). ( -32.90) >consensus GGAGCUGCGGCUCCUGUGAAAGGGCAAACAAAGCCCACUGACUGCUAAAUGCGGUCUAAAUGAAGUGUAACUACAAUUAGUAA_UGGGCUUUA (((((....))))).......((((.......))))...((((((.....))))))....((((((.((((((....))))...)).)))))) (-23.28 = -22.62 + -0.66)


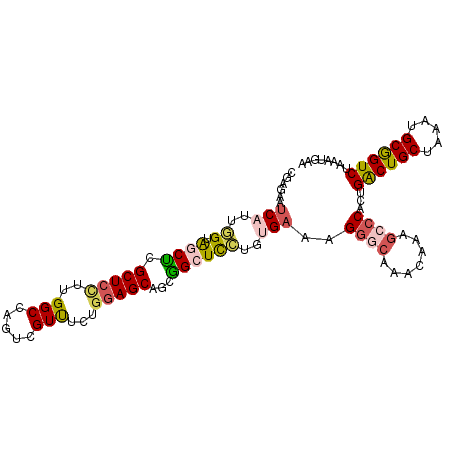
| Location | 7,704,780 – 7,704,884 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.95 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -24.55 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7704780 104 - 27905053 CGAGAAUCAUUGGUAGCUCGCUCCUUGGCCAGUCGUUGCUGGAGCCUCGGCUCCUUUGAAAGGGCAAACAAAGCCCACUGACUGCUGAAUGCGGUCUAAAUGAG ......(((((..(((..(((...(..((.((((......(((((....))))).......((((.......))))...))))))..)..)))..)))))))). ( -36.90) >DroVir_CAF1 5716 104 - 1 UGAGAUUCAGCCGCAGCACGCUCAUGGGCGAUACGUCGUUGUAGCAGCUGAUUGAGCGACAGGAGAUACAAAGCCCACUGACUGCUGAAGGCGGUCUGCAGGGA ((((((...(((.(((((.(.((((((((.....(((.((((....(((.....))).))))..))).....))))).)))))))))..))).)))).)).... ( -33.70) >DroSec_CAF1 1722 104 - 1 CGAGAAUCAUUGGUAGCUCGCUCCUUGGCCAGUCGUUUCUGGAGCGUCGGCUCCUUUGAAAGGGCAAACAAAGCCCACUGACUGCUAAAUGCGGUCUGUAUGAA ......(((..((.((((((((((..(((.....)))...))))))..))))))..)))..((((.......))))((.((((((.....)))))).))..... ( -39.20) >DroSim_CAF1 1718 104 - 1 CGAGAAUCAUUGGUAGCUCGCUCCUUGGCCAGUCGUUUCUGGAGCGUCGGCUCCUUUGAAAGGGCAAACAAAGCCCACUGACUGCUAAAUGCGGUCUGUAUGAG ......(((..((.((((((((((..(((.....)))...))))))..))))))..)))..((((.......))))((.((((((.....)))))).))..... ( -39.20) >DroYak_CAF1 1770 104 - 1 CGAGAAUCAUUGGUGGCUCGCUCUUUGGCCAGCCGUUUCUGGAGCUGGGGCUCCUGUGAAAGGGCAAAUAAUGCCCACUGACUGCUAAACGCGGUCUAAACGAA ((((..((((.((.(.((((((((.(((....))).....))))).))).).)).))))..(((((.....))))).))((((((.....))))))....)).. ( -38.20) >DroAna_CAF1 1434 104 - 1 UGGGAGCCAUGAGAAGCGCGCUCUCGGGCCAGCCGUUUCUGGAGCAGCUGAUUCUGGGACAGCACAAACAAUGCCCACUGGCUGCUAAAGGCAGUCUAGUGGAA ......(((.((..(((..((((((((.....))).....))))).)))..)).)))....(((.......)))(((((((((((.....)))).))))))).. ( -36.20) >consensus CGAGAAUCAUUGGUAGCUCGCUCCUUGGCCAGUCGUUUCUGGAGCAGCGGCUCCUGUGAAAGGGCAAACAAAGCCCACUGACUGCUAAAUGCGGUCUAAAUGAA ......(((..((.((((.(((((..(((.....)))...)))))...))))))..)))..((((.......))))...((((((.....))))))........ (-24.55 = -24.92 + 0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:55 2006