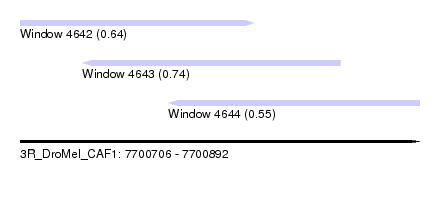
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,700,706 – 7,700,892 |
| Length | 186 |
| Max. P | 0.739826 |

| Location | 7,700,706 – 7,700,815 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.40 |
| Mean single sequence MFE | -28.44 |
| Consensus MFE | -24.25 |
| Energy contribution | -23.83 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
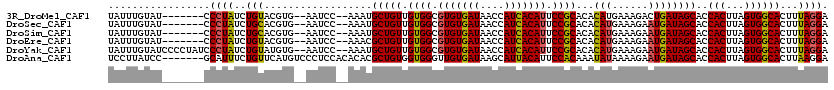
>3R_DroMel_CAF1 7700706 109 + 27905053 UAUUUGUAU-------CCCUAUCUGUACGUG--AAUCC--AAAUGCUGUUGUGGCGUGUGAUAACCAUCACAUUCCGCACACAUGAAAGACUGAUAGCACCACUUAGUGGCACUUUAGGA .........-------.((((...((.....--.....--...(((((((((((.(((((((....))))))).)))).(....).......)))))))((((...)))).))..)))). ( -26.40) >DroSec_CAF1 50403 109 + 1 UAUUUGUAU-------CCCUAUCUGCACGUG--AAUCC--AAAUGCUGUUGUGGCGUGUGAUAACCAUCACAUUCCGCACACAUGAAAGAAUGAUAGCACCACUUAGUGGCACUUUAGGA .........-------.((((..(((.....--.....--...(((((((((((.(((((((....))))))).)))))..(((......))))))))).(((...))))))...)))). ( -28.80) >DroSim_CAF1 50571 109 + 1 UAUUUGUAU-------CCCUAUCUGCACGUG--AAUCC--AAAUGCUGUUGUGGCGUGUGAUAACCAUCACAUUCCGCACACAUGAAAGAAUGAUAGCACCACUUAGUGGCACUUUAGGA .........-------.((((..(((.....--.....--...(((((((((((.(((((((....))))))).)))))..(((......))))))))).(((...))))))...)))). ( -28.80) >DroEre_CAF1 50974 109 + 1 UAUUUGUAU-------CCCUAUCUGUACGUG--AAUCC--AAACGCUGUUGUGGCGUGUGAUAACCAUCACAUUCCGCACACAUGAAAGAAUGAUAGCACCACUUAGUGGCACUUUAGGA .........-------.((((...((.....--.....--....((((((((((.(((((((....))))))).)))))..(((......)))))))).((((...)))).))..)))). ( -27.90) >DroYak_CAF1 52161 116 + 1 UAUUUGUAUCCCCUAUCCCUAUCUGUAUGUG--AAUCC--AAAUGCUGUUGUGGCGUGUGAUAACCAUCACAUUCCGCACACAUGAAAGAAUGAUAGCACCACUUAGUGGCACUUUAGGA ...........((((...(((((.((((.((--....)--).))))((((((((.(((((((....))))))).))))).))).........)))))..((((...)))).....)))). ( -28.70) >DroAna_CAF1 52482 113 + 1 UCCUUAUCC-------GCAUUUCUGUUCAUGUCCCUCCACACACGCUGUGGUGGGUUGUGAUAAGCAUUACAUUCCACAAAUAUAAAAGAAUGAUAGCACCACUUAGUGGCACUUAAGGA ((((((.((-------((........(((((.(((.(((((.....))))).))).)))))...((....(((((.............)))))...))........))))....)))))) ( -30.02) >consensus UAUUUGUAU_______CCCUAUCUGUACGUG__AAUCC__AAAUGCUGUUGUGGCGUGUGAUAACCAUCACAUUCCGCACACAUGAAAGAAUGAUAGCACCACUUAGUGGCACUUUAGGA .................((((..(((..................(((((.((((.(((((((....))))))).))))...(((......))))))))..(((...))))))...)))). (-24.25 = -23.83 + -0.41)

| Location | 7,700,735 – 7,700,855 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -32.65 |
| Consensus MFE | -28.56 |
| Energy contribution | -28.83 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7700735 120 - 27905053 UGAGCGAAACGUAAGCGGAAGUCCUUACAGCUUUUGCAUAUCCUAAAGUGCCACUAAGUGGUGCUAUCAGUCUUUCAUGUGUGCGGAAUGUGAUGGUUAUCACACGCCACAACAGCAUUU ...((((((.....((((((((.......)))))))).........((..((((...))))..)).......)))).((((.(((...((((((....)))))))))))))...)).... ( -35.10) >DroSec_CAF1 50432 120 - 1 UGAGCGAAACGUAAGCGGAAGUCCUUACAGCUUUUGCAUAUCCUAAAGUGCCACUAAGUGGUGCUAUCAUUCUUUCAUGUGUGCGGAAUGUGAUGGUUAUCACACGCCACAACAGCAUUU ...((((((.....((((((((.......)))))))).........((..((((...))))..)).......)))).((((.(((...((((((....)))))))))))))...)).... ( -35.10) >DroSim_CAF1 50600 120 - 1 UGAGCGAAACGUAAGCGGAAGUCCUUACAGCUUUUGCAUAUCCUAAAGUGCCACUAAGUGGUGCUAUCAUUCUUUCAUGUGUGCGGAAUGUGAUGGUUAUCACACGCCACAACAGCAUUU ...((((((.....((((((((.......)))))))).........((..((((...))))..)).......)))).((((.(((...((((((....)))))))))))))...)).... ( -35.10) >DroEre_CAF1 51003 108 - 1 ------------AAGCGGAAGUCCUUACAGCUUUUGCAUAUCCUAAAGUGCCACUAAGUGGUGCUAUCAUUCUUUCAUGUGUGCGGAAUGUGAUGGUUAUCACACGCCACAACAGCGUUU ------------..((((((((.......)))))))).........((..((((...))))..))............((((.(((...((((((....)))))))))))))......... ( -30.80) >DroYak_CAF1 52197 120 - 1 UGAGCGAAACGUAAGCGGAAGUCCUUACAGCUUUUGCAUAUCCUAAAGUGCCACUAAGUGGUGCUAUCAUUCUUUCAUGUGUGCGGAAUGUGAUGGUUAUCACACGCCACAACAGCAUUU ...((((((.....((((((((.......)))))))).........((..((((...))))..)).......)))).((((.(((...((((((....)))))))))))))...)).... ( -35.10) >DroAna_CAF1 52515 120 - 1 UAAACGAACCGUAAGCGGAAGUCCUUACAGCUUUUGCAUAUCCUUAAGUGCCACUAAGUGGUGCUAUCAUUCUUUUAUAUUUGUGGAAUGUAAUGCUUAUCACAACCCACCACAGCGUGU ...(((....((((((((((((.......)))))))).........((..((((...))))..)).........))))..((((((...((..((.....))..))...))))))))).. ( -24.70) >consensus UGAGCGAAACGUAAGCGGAAGUCCUUACAGCUUUUGCAUAUCCUAAAGUGCCACUAAGUGGUGCUAUCAUUCUUUCAUGUGUGCGGAAUGUGAUGGUUAUCACACGCCACAACAGCAUUU ..............((((((((.......)))))))).........((..((((...))))..))...........((((.((.((..((((((....))))))..)).))...)))).. (-28.56 = -28.83 + 0.28)

| Location | 7,700,775 – 7,700,892 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.40 |
| Mean single sequence MFE | -30.46 |
| Consensus MFE | -20.54 |
| Energy contribution | -22.93 |
| Covariance contribution | 2.39 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7700775 117 - 27905053 GAAGUUCAGGUUUACUCCAACUU-UU--UUUCGAGCAUCCUGAGCGAAACGUAAGCGGAAGUCCUUACAGCUUUUGCAUAUCCUAAAGUGCCACUAAGUGGUGCUAUCAGUCUUUCAUGU ...(((((((....(((.((...-..--.)).)))...)))))))((((.....((((((((.......)))))))).........((..((((...))))..)).......)))).... ( -31.40) >DroSec_CAF1 50472 119 - 1 GAAGUUCAGGUUUACUCCAACUU-UUUUUUUGGAGCAUCCUGAGCGAAACGUAAGCGGAAGUCCUUACAGCUUUUGCAUAUCCUAAAGUGCCACUAAGUGGUGCUAUCAUUCUUUCAUGU ...(((((((....((((((...-.....))))))...)))))))((((.....((((((((.......)))))))).........((..((((...))))..)).......)))).... ( -37.70) >DroSim_CAF1 50640 119 - 1 GAACUUCAGGUUUACUCCAACUU-UUUUUUUGGAGCAUCCUGAGCGAAACGUAAGCGGAAGUCCUUACAGCUUUUGCAUAUCCUAAAGUGCCACUAAGUGGUGCUAUCAUUCUUUCAUGU (((..(((((....((((((...-.....))))))...)))))(((((((((((((....)..))))).).)))))).........((..((((...))))..))........))).... ( -33.50) >DroEre_CAF1 51043 96 - 1 GAACUUUAGGUUUACUCCAACUC-GG---UUG--------------------AAGCGGAAGUCCUUACAGCUUUUGCAUAUCCUAAAGUGCCACUAAGUGGUGCUAUCAUUCUUUCAUGU ((((((((((.......((((..-.)---)))--------------------..((((((((.......))))))))....))))))))(((((...)))))....))............ ( -23.50) >DroYak_CAF1 52237 116 - 1 GAACUUCAGGCUUACUCCAACUC-GG---CUGGAGCAUCCUGAGCGAAACGUAAGCGGAAGUCCUUACAGCUUUUGCAUAUCCUAAAGUGCCACUAAGUGGUGCUAUCAUUCUUUCAUGU (((..(((((....(((((....-..---.)))))...)))))(((((((((((((....)..))))).).)))))).........((..((((...))))..))........))).... ( -31.90) >DroAna_CAF1 52555 113 - 1 ------CCUGGGU-CCUCAACUGACUCUAUUGGAGUUUCCUAAACGAACCGUAAGCGGAAGUCCUUACAGCUUUUGCAUAUCCUUAAGUGCCACUAAGUGGUGCUAUCAUUCUUUUAUAU ------...((.(-(.......(((((.....)))))........)).))....((((((((.......)))))))).........((..((((...))))..))............... ( -24.76) >consensus GAACUUCAGGUUUACUCCAACUU_UU__UUUGGAGCAUCCUGAGCGAAACGUAAGCGGAAGUCCUUACAGCUUUUGCAUAUCCUAAAGUGCCACUAAGUGGUGCUAUCAUUCUUUCAUGU .....(((((....((((((.........))))))...)))))...........((((((((.......)))))))).........((..((((...))))..))............... (-20.54 = -22.93 + 2.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:51 2006