
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,685,504 – 7,685,623 |
| Length | 119 |
| Max. P | 0.557457 |

| Location | 7,685,504 – 7,685,623 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.82 |
| Mean single sequence MFE | -40.98 |
| Consensus MFE | -25.76 |
| Energy contribution | -27.23 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7685504 119 + 27905053 UGAAUGCCAUCUAAUGAUCGAUUGAUCACCUCCUUUGUAUGUAUAACAGGGCAACGUCCAGUCCAUCUAUGGACCGGGUCCGCCUCCUUUUCGAGGAGUGGGCGGCCGGGAGGA-GCAGC ....(((.......(((((....)))))((((((((((.......))))(((.....((.(((((....))))).))((((((.(((((...))))))))))).))))))))).-))).. ( -45.00) >DroSec_CAF1 35360 116 + 1 UCAAUGCCAUCUAAUGAUCGAUUGAUCAUCUCCUUUG----UAUAACAGGGCAACGUCCAGUCCAUCUAUGUACCAUGUCCGCCUCCUUUUCGAGGAGUGGGCGGCCGGGAGCAGGCAGC ....((((.....((((((....))))))(((((..(----((((...((((........)))).....)))))..(((((((.(((((...))))))))))))...)))))..)))).. ( -37.20) >DroSim_CAF1 35363 116 + 1 ACAAUGCCAUCUAAUGAUCGAUUGAUCAUCUCCUUUG----UAUAACAGGGCAACGUCCAGUCCAUCUAUGGACAAGGUCCGCCUCCUUUUCGAGGAGUGGGCGGCCGGGAGGAGGCAGC ....((((......(((((....)))))((((((...----.......((((...)))).(((((....)))))..((.(((((..((((....))))..))))))))))))).)))).. ( -45.30) >DroEre_CAF1 35936 115 + 1 CCAAUGCCAUCUUAUGAUCGAUUGAUCAGCUCGUUUG----UAUACCAGGGCAACGUCCAGUCCAUCUAUGGACCAGGUCCGCCUCCUUUUCGAGGAGUGGGCGGACGGGAGGA-GCAGC ....((((.(((..(((((....)))))((((.....----.......))))..(((((.(((((....)))))...((((((.(((((...))))))))))))))))))).).-))).. ( -41.80) >DroYak_CAF1 35744 115 + 1 UCAAUGCCAUCUAACGAUCGAUUGAUCACCUCCUUUG----UAGAACAGGGCAACGUCCAGUCCAUCUAUGGACCAGGUCCGCCUCCUUUUCGAGGAGUGGGCGGACGGGAGGA-GCAGC ....(((........((((....)))).(((((.(((----(........))))(((((.(((((....)))))...((((((.(((((...))))))))))))))))))))).-))).. ( -45.20) >DroAna_CAF1 37910 99 + 1 UAAGGGUCAAACACCGAUCAGUUGGUCAUUCCGAGCC----AUUAGCAGGGCAACGUCCAGUCCAUCUGCGGA----GUCCAC--------CGAGCAGUGGGCGGCA-----UAGGAUGC ...((........))((((....))))..(((..(((----.......((((...)))).(((((.((((((.----.....)--------)..)))))))))))).-----..)))... ( -31.40) >consensus UCAAUGCCAUCUAAUGAUCGAUUGAUCAUCUCCUUUG____UAUAACAGGGCAACGUCCAGUCCAUCUAUGGACCAGGUCCGCCUCCUUUUCGAGGAGUGGGCGGCCGGGAGGA_GCAGC ....(((........((((....))))..(((((..............((((...)))).((((......))))...((((((.((((.....))))))))))....)))))...))).. (-25.76 = -27.23 + 1.48)

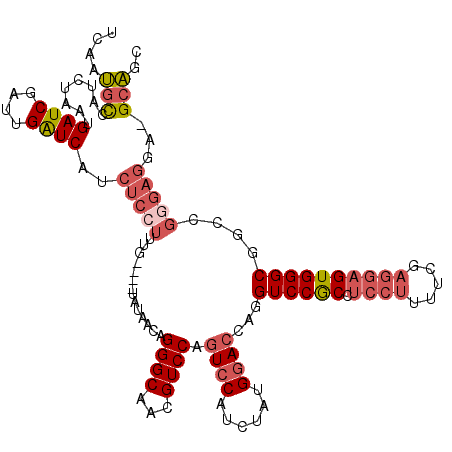
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:34 2006