
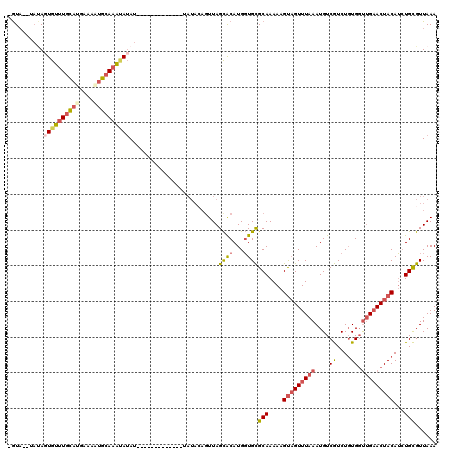
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,679,494 – 7,679,632 |
| Length | 138 |
| Max. P | 0.999884 |

| Location | 7,679,494 – 7,679,600 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.26 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -14.30 |
| Energy contribution | -15.78 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7679494 106 - 27905053 -GUACGUAUAUUGUUUGUAUGAAAAUGCAAAUAUAU-------------UAUACAGUUAGCACAUGGUGCGCAAAAAGUAGUUUAAAUGUCGUCUGUGGUUGAACUACAUCUGCGUUAAA -....(((((.(((((((((....)))))))))...-------------))))).....((((...))))(((....(((((((((....(....)...)))))))))...)))...... ( -27.20) >DroSec_CAF1 29432 103 - 1 -GUA--UAUAGUGU-UGCAUGAAAGUGCAAAUAUAU-------------UAUACAGUUAGCACAUGGUGCGCAAAAAGUAGUUUAAAUGUCGUCUGUGGUUGAACUACAUCUGCGUUAAA -(((--((((...(-(((((....))))))...)).-------------))))).....((((...))))(((....(((((((((....(....)...)))))))))...)))...... ( -25.90) >DroSim_CAF1 29440 104 - 1 -GUA--UAUAGUGUUUGCAUGAAAAUGCAAAUAUAU-------------UAUACAUUUAGCACAUGGUGCGCAAAAAGUAGUUUAAAUGUCGUCUGUGGUUGAACUACAUCUGCGUUAAA -(((--((..((((((((((....))))))))))..-------------))))).....((((...))))(((....(((((((((....(....)...)))))))))...)))...... ( -30.30) >DroYak_CAF1 29571 114 - 1 -AUC-----AGUGUUUGCAUGAAUUUGGAAAAAUAUACAUACUCGUAUAUAUAAAGUUAGCACAUGGUGCGCAAAAAGUAGUUUAAAUGUCGUCUGUGGUUGAACUACAUGUGCGUUAAA -(((-----((((((.((......((....))((((((......)))))).....)).))))).))))(((((....(((((((((....(....)...)))))))))...))))).... ( -25.00) >DroAna_CAF1 32211 96 - 1 AGUA--CACA-UAUUUGC---AAAUUGCAAGUAAAU------------------CGUUAGUGCAUGUUGUGCAAAAAGCUGUUUAAAUGUCAUCUCUGUCCGCACUACAACUGUGUUAAA ...(--((((-(((((((---.....)))))))...------------------.((.(((((.......((.....)).........(.((....)).).)))))))...))))).... ( -18.20) >consensus _GUA__UAUAGUGUUUGCAUGAAAAUGCAAAUAUAU_____________UAUACAGUUAGCACAUGGUGCGCAAAAAGUAGUUUAAAUGUCGUCUGUGGUUGAACUACAUCUGCGUUAAA ..........((((((((((....)))))))))).........................((((...))))(((....(((((((((....((....)).)))))))))...)))...... (-14.30 = -15.78 + 1.48)


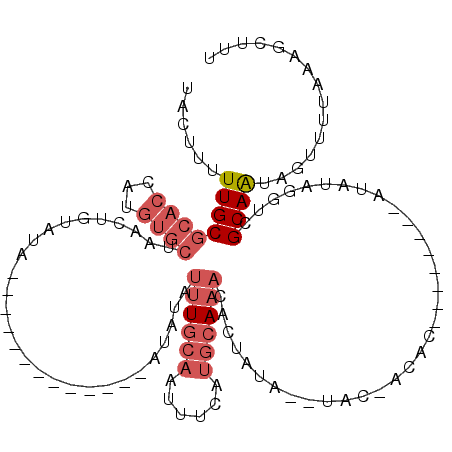
| Location | 7,679,534 – 7,679,632 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.51 |
| Mean single sequence MFE | -18.94 |
| Consensus MFE | -4.88 |
| Energy contribution | -6.52 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.26 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7679534 98 + 27905053 UACUUUUUGCGCACCAUGUGCUAACUGUAUA-------------AUAUAUUUGCAUUUUCAUACAAACAAUAUACGUAC-AUAU--------AUGUAGGUCGCAAUAGUUUUAAAGCUUU .(((..(((((.(((.(((((.....)))))-------------............................((((((.-...)--------))))))))))))).)))........... ( -17.20) >DroSec_CAF1 29472 89 + 1 UACUUUUUGCGCACCAUGUGCUAACUGUAUA-------------AUAUAUUUGCACUUUCAUGCA-ACACUAUA--UAC-ACAC--------A------UCGCAAUAGUUUUAAAGCUUU .(((..(((((....(((((.....((((((-------------......(((((......))))-).....))--)))-))))--------)------)))))).)))........... ( -15.80) >DroSim_CAF1 29480 96 + 1 UACUUUUUGCGCACCAUGUGCUAAAUGUAUA-------------AUAUAUUUGCAUUUUCAUGCAAACACUAUA--UAC-ACAC--------AUAUAGGUCGCAAUAGUUUUAAAGCUUU .(((..(((((.((((((((.....((((((-------------.....(((((((....))))))).....))--)))-)..)--------)))).)))))))).)))........... ( -22.90) >DroYak_CAF1 29611 104 + 1 UACUUUUUGCGCACCAUGUGCUAACUUUAUAUAUACGAGUAUGUAUAUUUUUCCAAAUUCAUGCAAACACU-----GAU-A--U--------AUGUAGGUCGCAAUAGUUUUAAAGCUUU .(((..(((((.(((..((....))..((((((((..(((.(((((((((....))))..)))))...)))-----..)-)--)--------))))))))))))).)))........... ( -18.40) >DroAna_CAF1 32251 94 + 1 AGCUUUUUGCACAACAUGCACUAACG------------------AUUUACUUGCAAUUU---GCAAAUA-UGUG--UACUACACAGCGUUGUCUACAAAUAGCAG--CUAUUGAAGCUUU .(((.((((.(((((..((.......------------------......((((.....---))))...-((((--.....)))))))))))...)))).)))((--((.....)))).. ( -20.40) >consensus UACUUUUUGCGCACCAUGUGCUAACUGUAUA_____________AUAUAUUUGCAAUUUCAUGCAAACACUAUA__UAC_ACAC________AUAUAGGUCGCAAUAGUUUUAAAGCUUU ......((((((((...))))............................((((((......))))))..................................))))............... ( -4.88 = -6.52 + 1.64)

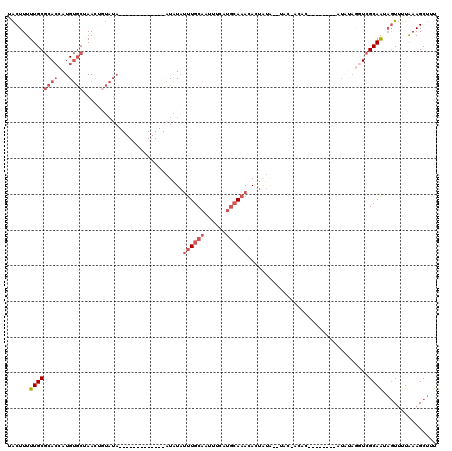
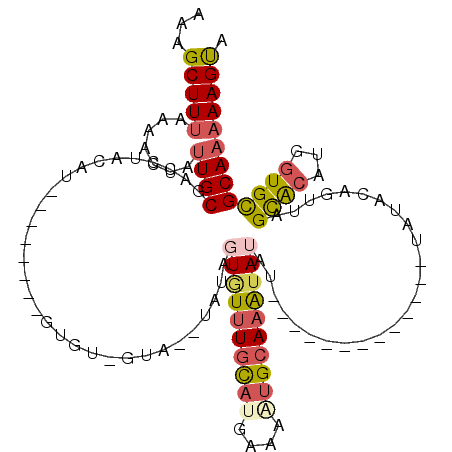
| Location | 7,679,534 – 7,679,632 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.51 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -12.39 |
| Energy contribution | -13.43 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.50 |
| SVM decision value | 4.38 |
| SVM RNA-class probability | 0.999884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7679534 98 - 27905053 AAAGCUUUAAAACUAUUGCGACCUACAU--------AUAU-GUACGUAUAUUGUUUGUAUGAAAAUGCAAAUAUAU-------------UAUACAGUUAGCACAUGGUGCGCAAAAAGUA ...(((((.......((((((((((((.--------...)-))).(((((.(((((((((....)))))))))...-------------)))))...........))).)))))))))). ( -24.61) >DroSec_CAF1 29472 89 - 1 AAAGCUUUAAAACUAUUGCGA------U--------GUGU-GUA--UAUAGUGU-UGCAUGAAAGUGCAAAUAUAU-------------UAUACAGUUAGCACAUGGUGCGCAAAAAGUA ...(((((...((((((((..------(--------((((-(.(--((((...(-(((((....)))))).)))))-------------))))))....))).)))))......))))). ( -25.40) >DroSim_CAF1 29480 96 - 1 AAAGCUUUAAAACUAUUGCGACCUAUAU--------GUGU-GUA--UAUAGUGUUUGCAUGAAAAUGCAAAUAUAU-------------UAUACAUUUAGCACAUGGUGCGCAAAAAGUA ...(((((.......(((((..((((.(--------((((-(((--((..((((((((((....))))))))))..-------------)))))))...))).))))..)))))))))). ( -31.41) >DroYak_CAF1 29611 104 - 1 AAAGCUUUAAAACUAUUGCGACCUACAU--------A--U-AUC-----AGUGUUUGCAUGAAUUUGGAAAAAUAUACAUACUCGUAUAUAUAAAGUUAGCACAUGGUGCGCAAAAAGUA ...(((((.......((((((((.((((--------.--.-...-----.)))).(((.(((.((((.....((((((......)))))).)))).))))))...))).)))))))))). ( -17.61) >DroAna_CAF1 32251 94 - 1 AAAGCUUCAAUAG--CUGCUAUUUGUAGACAACGCUGUGUAGUA--CACA-UAUUUGC---AAAUUGCAAGUAAAU------------------CGUUAGUGCAUGUUGUGCAAAAAGCU ..((((.....))--))(((.((((((((((.(((((((.....--))).-(((((((---.....)))))))...------------------....))))..)))).)))))).))). ( -25.80) >consensus AAAGCUUUAAAACUAUUGCGACCUACAU________GUGU_GUA__UAUAGUGUUUGCAUGAAAAUGCAAAUAUAU_____________UAUACAGUUAGCACAUGGUGCGCAAAAAGUA ...(((((.......((((...............................((((((((((....)))))))))).........................((((...))))))))))))). (-12.39 = -13.43 + 1.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:26 2006