
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,661,390 – 7,661,547 |
| Length | 157 |
| Max. P | 0.942005 |

| Location | 7,661,390 – 7,661,510 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -43.45 |
| Consensus MFE | -37.74 |
| Energy contribution | -38.10 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7661390 120 - 27905053 CAGGCGGGCAAUUGCCGAAAUGAAUGAAGAUGAAGUGAACCGGGCUUGUCCGGCACUUGAACUGCCUGCCAGCACUCCGGUCCCAAGAUUGAUUGGCCUCCAGUCGAAGUCGACUCGGAU ..(((((((.(((.....)))......((.(.(((((..(((((....)))))))))).).))))))))).....(((((((......((((((((...))))))))....))).)))). ( -42.00) >DroSec_CAF1 16847 120 - 1 CAGGCGGGCAAUUACCCAAAUGAAUGAAGAUGAAGUGAACCGGGCUUGUCCGGCACUUGAACUGCCUGCCAGCACUCCGGUCCCAAGAUUGAUUGGCCUCCAGUCGAAGUCGCCUCGGAU ..(((((((.......((......)).((.(.(((((..(((((....)))))))))).).))))))))).....(((((......((((((((((...)))))))..)))...))))). ( -38.90) >DroSim_CAF1 16426 120 - 1 CAGGCGGGCAAUUGCCCAAAUGAAUGAAGAUGAAGUGAACCGGGCUUGUCCGGCACUUGAACUGCCUGCCAGCACUCCGGUCCCAAGAUUGAUUGGCCUCCAGCCGAAGUCGCCUCGGAU (((((((((....))))..........((.(.(((((..(((((....)))))))))).).))))))).......(((((......((((..(((((.....)))))))))...))))). ( -41.10) >DroEre_CAF1 16748 120 - 1 CAGGCGGGCAAUUGCCCAAAUGAAUGAAGAUGAAGUGAACCGGGCUUGUCCGGCACUUGAACUGCCUGCUAGCACUCCGGGCCCAAGAUUGAUUGGCCUCCAGUCGAAGUCGACUGGGAU (((((((((....))))..........((.(.(((((..(((((....)))))))))).).)))))))..........(((((.((......)))))))(((((((....)))))))... ( -48.70) >DroYak_CAF1 15923 120 - 1 CAGGCGGGCAAUUGCCCAAAUGAAUGAAGAUGAAGUGAACCGGGCUUGUCCGGCACUUGAACUGCCUGCCAGCACUCCGGGCCCAAGAUUGAUUGGCCUCCAGUCGAAGUCGACUCGGAU (((((((((....))))..........((.(.(((((..(((((....)))))))))).).))))))).......((((((((.((......))))))...(((((....))))))))). ( -46.10) >DroAna_CAF1 19731 120 - 1 CAGGCGGGCAAUUGCCCAAAUGAAUGAAGAUGAAGUGAAGCGGGCCUGCCCGGCACUUGAACUGCCUGCCAGAACUCCAGCCCCAAGAUUGAUUGGCCUCCGGUUGAAGUUGGCUCGGAA .((((((((....))))..........((.(.(((((...((((....)))).))))).).))))))((((..(((.(((((((((......)))).....))))).)))))))...... ( -43.90) >consensus CAGGCGGGCAAUUGCCCAAAUGAAUGAAGAUGAAGUGAACCGGGCUUGUCCGGCACUUGAACUGCCUGCCAGCACUCCGGUCCCAAGAUUGAUUGGCCUCCAGUCGAAGUCGACUCGGAU (((((((((....))))..........((.(.(((((..(((((....)))))))))).).))))))).......(((((..((((......))))...))(((((....))))).))). (-37.74 = -38.10 + 0.36)

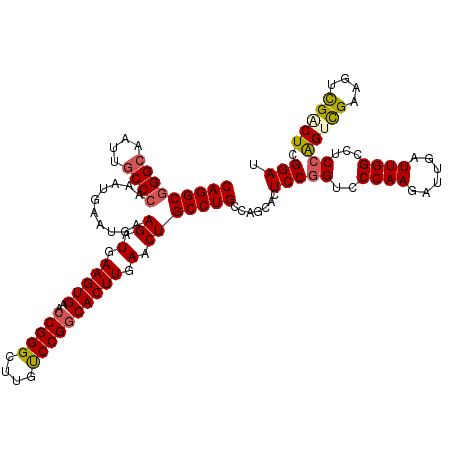
| Location | 7,661,430 – 7,661,547 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -38.37 |
| Consensus MFE | -34.48 |
| Energy contribution | -34.57 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7661430 117 + 27905053 CCGGAGUGCUGGCAGGCAGUUCAAGUGCCGGACAAGCCCGGUUCACUUCAUCUUCAUUCAUUUCGGCAAUUGCCCGCCUGUCGCCUGCACAAAUUUCCCAUCCCUG---GUCCUUUUCGC ..(((((((((((((((((...(((((((((......))))..)))))...))...........(((....))).))))))))...)))).......(((....))---))))....... ( -37.30) >DroSec_CAF1 16887 117 + 1 CCGGAGUGCUGGCAGGCAGUUCAAGUGCCGGACAAGCCCGGUUCACUUCAUCUUCAUUCAUUUGGGUAAUUGCCCGCCUGUCGCCUGCACACAGUUCCCAUCCCUG---GUCCUUUUCGC ..(((((((((((((((((...(((((((((......))))..)))))...))..........((((....))))))))))))...)))).......(((....))---))))....... ( -39.00) >DroSim_CAF1 16466 117 + 1 CCGGAGUGCUGGCAGGCAGUUCAAGUGCCGGACAAGCCCGGUUCACUUCAUCUUCAUUCAUUUGGGCAAUUGCCCGCCUGUCGCCUGCACACAGUUCCCAUUCCUG---GUCCUUUUCGC ..(((((((((((((((((...(((((((((......))))..)))))...))..........((((....))))))))))))...)))).......(((....))---))))....... ( -41.20) >DroEre_CAF1 16788 120 + 1 CCGGAGUGCUAGCAGGCAGUUCAAGUGCCGGACAAGCCCGGUUCACUUCAUCUUCAUUCAUUUGGGCAAUUGCCCGCCUGUCGCCUGCACAAAGUUCCCGUCCCUUUCCAUCCUUCGCGC ..(((((((..((((((((...(((((((((......))))..)))))...))..........((((....)))))))))).....))))((((.........))))...)))....... ( -35.80) >DroYak_CAF1 15963 120 + 1 CCGGAGUGCUGGCAGGCAGUUCAAGUGCCGGACAAGCCCGGUUCACUUCAUCUUCAUUCAUUUGGGCAAUUGCCCGCCUGUCGCCUGCACAAAGUACCCAUCCCUUUCCAUCCUGUUCGC ..((.(((((.((((((((...(((((((((......))))..)))))...)).....((..(((((....)))))..))..))))))....)))))))..................... ( -40.30) >DroAna_CAF1 19771 107 + 1 CUGGAGUUCUGGCAGGCAGUUCAAGUGCCGGGCAGGCCCGCUUCACUUCAUCUUCAUUCAUUUGGGCAAUUGCCCGCCUGUCGCCCGAACAGUGU--CCUUUC-----------UUUCGC ..(((((((((((((((((...(((((.((((....))))...)))))...))..........((((....))))))))))))...))))....)--))....-----------...... ( -36.60) >consensus CCGGAGUGCUGGCAGGCAGUUCAAGUGCCGGACAAGCCCGGUUCACUUCAUCUUCAUUCAUUUGGGCAAUUGCCCGCCUGUCGCCUGCACAAAGUUCCCAUCCCUG___GUCCUUUUCGC .....((((((((((((((...(((((((((......))))..)))))...))..........((((....))))))))))))...)))).............................. (-34.48 = -34.57 + 0.09)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:15 2006