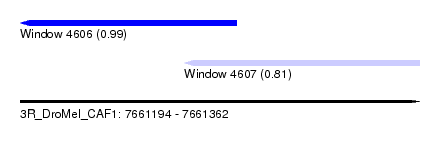
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,661,194 – 7,661,362 |
| Length | 168 |
| Max. P | 0.994720 |

| Location | 7,661,194 – 7,661,285 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -21.36 |
| Consensus MFE | -18.08 |
| Energy contribution | -18.44 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7661194 91 - 27905053 ACG--GGUGGCGCAUCUCAAGUCUCUGGAGUGCAAUCAAAGUGCAACAAUAGAGGAAAGCGGAAAGUGUUUGAGAUGAGAAAAGAAAGAUGAU ...--.....(.(((((((((((((((...((((.......))))....))))))...((.....)).))))))))).).............. ( -20.10) >DroSec_CAF1 16645 93 - 1 ACGAGGGUGGCGCAUCUCAAGUCUCUGGAGUGCAAUCAAAGUGCAACAAUAGAGGAAAGCGAAAAGUGUUUGAGAUGGGAAAAGAUAGAUGAA ..........(.(((((((((((((((...((((.......))))....))))))...((.....)).))))))))).).............. ( -21.70) >DroSim_CAF1 16225 93 - 1 ACGAGGGUGGCGCAUCUCAAGUCUCUGGAGUGCAAUCAAAGUGCAACAAUAGAGGAAAGCGAAAAGUGUUUGAGAUGGGAAAAGAUAGAUGAA ..........(.(((((((((((((((...((((.......))))....))))))...((.....)).))))))))).).............. ( -21.70) >DroEre_CAF1 16552 91 - 1 ACG--GGUAGCGAGUCUCAAGCCUCUGGAGUGCAAUCAAAGUGCAACAAUAGAGGACAGGGAAAUGCGUUUGAAAUGAGAAAAGAUAGAUGAA ...--....(((..((((...((((((...((((.......))))....))))))...))))..))).......................... ( -21.30) >DroYak_CAF1 15727 91 - 1 ACG--GGUAGCGCAUCUCAAGCCUCUGGAGUGCAAUCAAAGAGCAACAAUAGAGGAAAGGGAAUUGCGUUUGAGAUGAGAAAAGAUAGAUGAA .((--...((((((((((...((((((...(((.........)))....))))))...))))..)))))).....))................ ( -22.00) >consensus ACG__GGUGGCGCAUCUCAAGUCUCUGGAGUGCAAUCAAAGUGCAACAAUAGAGGAAAGCGAAAAGUGUUUGAGAUGAGAAAAGAUAGAUGAA ..........(.(((((((((((((((...((((.......))))....))))))...((.....)).))))))))).).............. (-18.08 = -18.44 + 0.36)

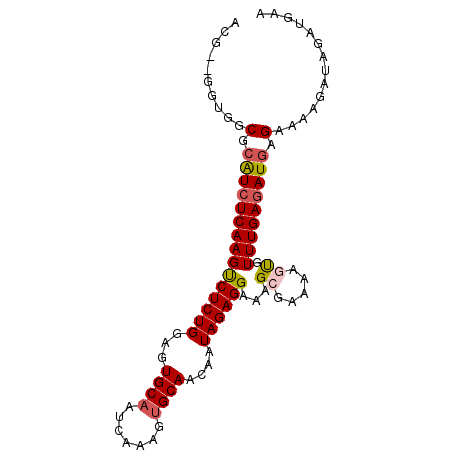
| Location | 7,661,263 – 7,661,362 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.87 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -21.01 |
| Energy contribution | -21.27 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7661263 99 - 27905053 CACUUCACU---GUCCUCGGGCGACUUUUCGUCCUCUUUGCUUUCAAAUUAGUUGGCCUGUCAAAAACUUAACGAGGCCAACG--GGUGGCGCAUCUCAA----------------GUCU .........---((((((((((((....)))))).................((((((((((.((....)).)).)))))))))--)).))).........----------------.... ( -28.20) >DroSec_CAF1 16714 104 - 1 CCCCUCACUCGGGUCCUCGGGCGACUUUUCGUCCUUUUUGCUUUCAAAUUAGUUGGCCUGUCAAAAACUUAACGAGGCCAACGAGGGUGGCGCAUCUCAA----------------GUCU .(((......)))(((((((((((....)))))).................((((((((((.((....)).)).))))))))))))).(((.........----------------))). ( -33.50) >DroSim_CAF1 16294 104 - 1 CCCUUCACUCGGGUCCUCGGGCGACUUUUCGUCCUUUUUGCUUUCAAAUUAGUUGGCCUGUCAAAAACUUAACGAGGCCAACGAGGGUGGCGCAUCUCAA----------------GUCU (((.......)))(((((((((((....)))))).................((((((((((.((....)).)).))))))))))))).(((.........----------------))). ( -32.70) >DroEre_CAF1 16621 99 - 1 CCCUUCGCU---GUCCUCGGGCGACUUUUCGUCCUUUUUGCUUUCAAAUUAGUUGGCCUGUCAAAAACUUAACGAGGCCAACG--GGUAGCGAGUCUCAA----------------GCCU ...((((((---..((..((((((....)))))).................((((((((((.((....)).)).)))))))))--)..))))))......----------------.... ( -35.50) >DroYak_CAF1 15796 99 - 1 CCCUUCACU---GGCCUCCGGCGACUUUUCGUCCUCUCUGCUUUCAAAUUAGUUGGCCUGUCAAAAACUUAACGAGGCCAACG--GGUAGCGCAUCUCAA----------------GCCU .........---(((....(((((....)))))......(((.((......((((((((((.((....)).)).)))))))).--)).))).........----------------))). ( -25.20) >DroAna_CAF1 19578 114 - 1 AUUUUGCCU---GUCCUCAGGCCACUUUU-GUCCUUUUCGCACCGAAAUUAGUUGGCCUGUCAGAGAGUUAACGAGGCCAGCG--AGUGGCCGAAGUUGACUGCAAUGAUGGUGCUGACC .....((((---(....))))).......-.....(((((...)))))...((..((((((((..((((((((..(((((...--..)))))...))))))).)..)))))).))..)). ( -40.20) >consensus CCCUUCACU___GUCCUCGGGCGACUUUUCGUCCUUUUUGCUUUCAAAUUAGUUGGCCUGUCAAAAACUUAACGAGGCCAACG__GGUGGCGCAUCUCAA________________GUCU ..................((((((....)))))).....(((.((......((((((((((.((....)).)).))))))))...)).)))............................. (-21.01 = -21.27 + 0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:13 2006