| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,108,366 – 1,108,485 |
| Length | 119 |
| Max. P | 0.619233 |

| Location | 1,108,366 – 1,108,485 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.29 |
| Mean single sequence MFE | -46.82 |
| Consensus MFE | -26.29 |
| Energy contribution | -27.15 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
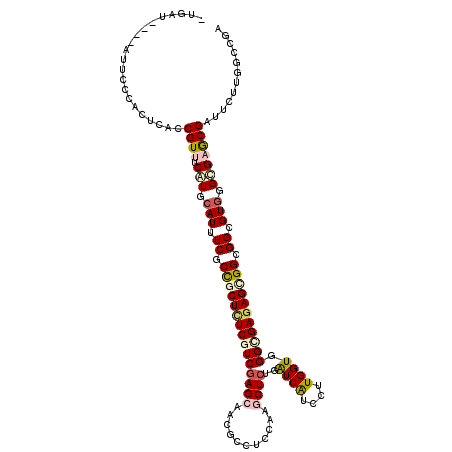
>3R_DroMel_CAF1 1108366 119 + 27905053 -CGAUCCUGACCGGCACUCACCGUUUCGUGCAUUUCGCCGCUCUCGUCGAGGAGAGCCUCCAGGCUCUGGUCGUCCUUGGUGGGCGAGAGAGGGGGCGUGGGCGCGCGGUUCUUGGCCGA -..........((((....((((...(((((....((((.(((((.((....((((((....))))))..((((((.....)))))))))))))))))...))))))))).....)))). ( -55.20) >DroVir_CAF1 35748 115 + 1 -UGCU----CUUCCCACUCACCGUUUCAUGCAUUUCGCCGCUUUCAUCUAGCAACGCCUCCAAGCUCUGAUCGUCUUUGGUUGGCGAGAGCGGCGGCGUGGGCGAGCGAUUCUUGGCCGU -.(((----(..(((((.....((.....))...(((((((((((.....(...)(((.(((((..(.....)..)))))..)))))))))))))).))))).))))............. ( -40.90) >DroGri_CAF1 48643 119 + 1 -UGAUUCUGUUUCCCACUCACCGUUUCAUGCAUUUCGCUGCUUUCGUCGAGCAACGCCUCCAAGCUCUGAUCAUCUUUGGUAGGCGAGAGCGGCGGCGUCGGUGAGCGAUUUUUCGCCGU -................((((((....((((....(((((((((((((((((...........))))..((((....)))).)))))))))))))))))))))))((((....))))... ( -44.20) >DroWil_CAF1 44449 110 + 1 ----------UCAAAACUCACCGUUUCAUGCAUUUCACCGCUCUCGUCGAGCAGUGCCUCCAGACUCUGAUCAUCCUUGGUGGGUGAUAGAGGCGGUGUGGGUGCACGAUUUUUGGCCGA ----------..........(((..((.(((((((((((((((.....))))...((((((((...)))(((((((.....))))))).))))))))).))))))).))....))).... ( -40.30) >DroMoj_CAF1 37631 115 + 1 -UGCU----CUACCCACUCACCGUUUCAUGCAUUUCACCGCUUUCGUCGAGCAAUGCCUCCAAGCUCUGAUCAUCUUUGGUGGGCGAGAGCGGCGGUGUGGGCGAGCGAUUCUUGACUGA -.(((----(..(((((..(((((.....))......(((((((((((((((...........))))..((((....)))).))))))))))).)))))))).))))............. ( -42.60) >DroAna_CAF1 40550 116 + 1 UCGAUC--GACUG--ACUCACCGUUUCGUGCAUUUCUCCGCUCUCAUCGAGCAGCGCCUCCAGGCUCUGGUCGUCCUUGGUGGGCGAGAGUGGGGGCGUGGGCGAGCGGUUUUUGGCGGC ....((--(.(..--(...((((((.(((.(((.(((((((((((...((.(((.(((....))).))).))((((.....))))))))))))))).))).)))))))))..)..)))). ( -57.70) >consensus _UGAU____AUUCCCACUCACCGUUUCAUGCAUUUCGCCGCUCUCGUCGAGCAACGCCUCCAAGCUCUGAUCAUCCUUGGUGGGCGAGAGCGGCGGCGUGGGCGAGCGAUUCUUGGCCGA .....................((((.(((.(((.((.(((((((((((((((...........))))..((((....)))).))))))))))).)).))).)))))))............ (-26.29 = -27.15 + 0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:28 2006