
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,606,062 – 7,606,209 |
| Length | 147 |
| Max. P | 0.918654 |

| Location | 7,606,062 – 7,606,169 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -26.05 |
| Energy contribution | -27.17 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7606062 107 - 27905053 CACAGGCGCUUAUGCAAGUGCACGUGUAUAUGUGAAUUAACACAUAAAAAUUAAGGUGCAGUCGAAUCGACUUGCCGGUCUCCUGCGCAUUUAUUU-UGUUACUUAAU------------ (((..((((((....))))))..)))..((((((......))))))(((((....((((((..((.(((......)))))..))))))....))))-)..........------------ ( -28.80) >DroSec_CAF1 1484 120 - 1 CACAGGCGUUUAUGCAAGUGCACGUGUAUGUGUGAAUUAACACAUAAAAAUUAAGGUGCAGUCGAAUCGACUUGCCGGUCUCCUGCGACUUUAUUUUUGCUAUUAAAGACAUCAGUUAUU ....((((..........(((((.(.(((((((......))))))).......).)))))((((...)))).)))).((((...((((........))))......)))).......... ( -27.40) >DroSim_CAF1 1486 120 - 1 CACAGGCGUUUAUGCAAGUGCACGUGUAUGUGUGAAUUAACACAUAAAAAUUAAGGUGCAGUCGAAUCGACUUGCCGGUCUCCUGCGCCUUUAUUUUUGCUACUAAAGACAUCAGUUAUU (((..(((....)))..)))...((((((((((......))))))((((((.(((((((((..((.(((......)))))..))))))))).))))))..........))))........ ( -32.50) >consensus CACAGGCGUUUAUGCAAGUGCACGUGUAUGUGUGAAUUAACACAUAAAAAUUAAGGUGCAGUCGAAUCGACUUGCCGGUCUCCUGCGCCUUUAUUUUUGCUACUAAAGACAUCAGUUAUU (((..((((((....))))))..)))(((((((......)))))))(((((.(((((((((..((.(((......)))))..))))))))).)))))....................... (-26.05 = -27.17 + 1.11)

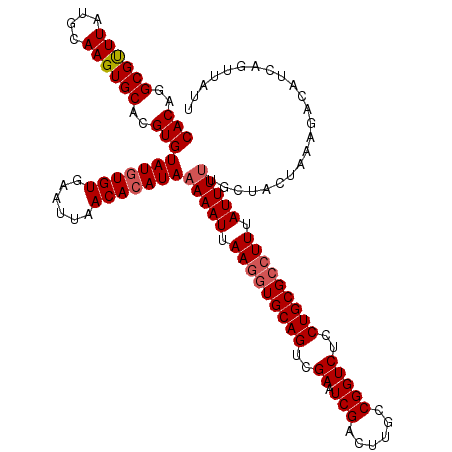

| Location | 7,606,089 – 7,606,209 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -27.79 |
| Energy contribution | -27.57 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7606089 120 + 27905053 GACCGGCAAGUCGAUUCGACUGCACCUUAAUUUUUAUGUGUUAAUUCACAUAUACACGUGCACUUGCAUAAGCGCCUGUGUGUGCAUGUCUGUAUGUCUUUUCUCAUUCUUGUUUUUUCC .....(((.((((...)))))))..............(((..((...(((((((.((((((((..(((((......))))))))))))).)))))))...))..)))............. ( -28.50) >DroSec_CAF1 1524 120 + 1 GACCGGCAAGUCGAUUCGACUGCACCUUAAUUUUUAUGUGUUAAUUCACACAUACACGUGCACUUGCAUAAACGCCUGUGUGUGCAUGUCUGUAUGUCUUUUCUCAUUCUUGUUUUUUCC ....((((((..((...(((((((..........(((((((......))))))).((((((((..(((((......))))))))))))).)))).)))...)).....))))))...... ( -28.80) >DroSim_CAF1 1526 120 + 1 GACCGGCAAGUCGAUUCGACUGCACCUUAAUUUUUAUGUGUUAAUUCACACAUACACGUGCACUUGCAUAAACGCCUGUGUGUGCAUGUCUGUAUGUCUUUCCUCAUUCUUGUUUUUUCC ....((((((..((...(((((((..........(((((((......))))))).((((((((..(((((......))))))))))))).)))).))).....))...))))))...... ( -28.20) >consensus GACCGGCAAGUCGAUUCGACUGCACCUUAAUUUUUAUGUGUUAAUUCACACAUACACGUGCACUUGCAUAAACGCCUGUGUGUGCAUGUCUGUAUGUCUUUUCUCAUUCUUGUUUUUUCC ....((((((..((...(((((((..........(((((((......))))))).((((((((..(((((......))))))))))))).)))).))).....))...))))))...... (-27.79 = -27.57 + -0.22)

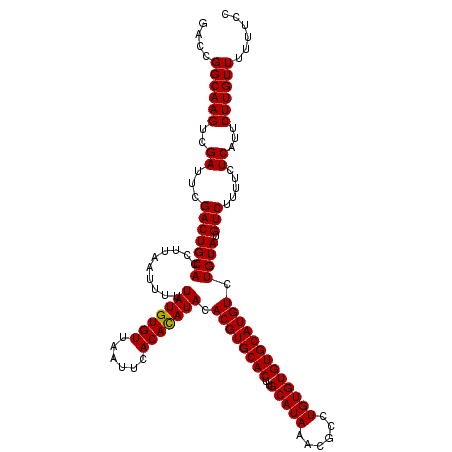

| Location | 7,606,089 – 7,606,209 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -27.31 |
| Energy contribution | -26.87 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7606089 120 - 27905053 GGAAAAAACAAGAAUGAGAAAAGACAUACAGACAUGCACACACAGGCGCUUAUGCAAGUGCACGUGUAUAUGUGAAUUAACACAUAAAAAUUAAGGUGCAGUCGAAUCGACUUGCCGGUC ........((((.(((........)))...(((.(((((((((..((((((....))))))..)))).((((((......)))))).........)))))))).......))))...... ( -30.40) >DroSec_CAF1 1524 120 - 1 GGAAAAAACAAGAAUGAGAAAAGACAUACAGACAUGCACACACAGGCGUUUAUGCAAGUGCACGUGUAUGUGUGAAUUAACACAUAAAAAUUAAGGUGCAGUCGAAUCGACUUGCCGGUC ......................(((...((.((((((((.(((..(((....)))..)))...)))))))).))....................((((.(((((...))))))))).))) ( -28.30) >DroSim_CAF1 1526 120 - 1 GGAAAAAACAAGAAUGAGGAAAGACAUACAGACAUGCACACACAGGCGUUUAUGCAAGUGCACGUGUAUGUGUGAAUUAACACAUAAAAAUUAAGGUGCAGUCGAAUCGACUUGCCGGUC ......................(((...((.((((((((.(((..(((....)))..)))...)))))))).))....................((((.(((((...))))))))).))) ( -28.30) >consensus GGAAAAAACAAGAAUGAGAAAAGACAUACAGACAUGCACACACAGGCGUUUAUGCAAGUGCACGUGUAUGUGUGAAUUAACACAUAAAAAUUAAGGUGCAGUCGAAUCGACUUGCCGGUC ........((((.(((........)))...(((.(((((((((..((((((....))))))..)))).((((((......)))))).........)))))))).......))))...... (-27.31 = -26.87 + -0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:56 2006