
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,605,529 – 7,605,689 |
| Length | 160 |
| Max. P | 0.932839 |

| Location | 7,605,529 – 7,605,649 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -45.73 |
| Consensus MFE | -43.57 |
| Energy contribution | -43.47 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
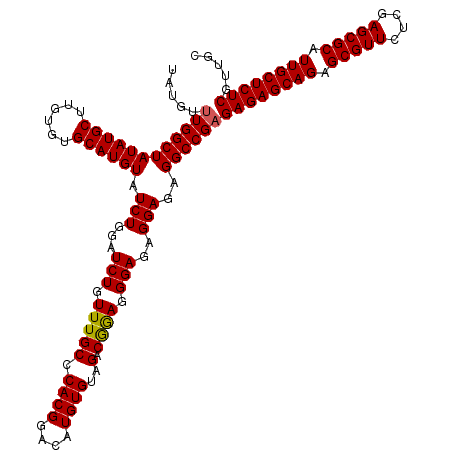
>3R_DroMel_CAF1 7605529 120 + 27905053 UAUGUUUGGCUAUAUGCUUGUGUGCAUGUAUCUGGAUCUGUUUGCCCACGGACAUGUGUAGACGAAGGGAAAGGAGAGGCCGAGAGAGCAGAGCGUUCUCGAGCGCAUUGCUCUCGUUGC .....((((((((((((......)))))).......(((.(((.(((.((.((....))...))..))).))).)))))))))((((((((.(((((....))))).))))))))..... ( -44.10) >DroSec_CAF1 945 119 + 1 UAUGU-UGGCUAUAUGCUUGUGUGCAUGUAUCUGGAUCUGUUUGCCCACGGACAUGUGUUGACGGAGGGAGAGGAGAGGCCGAGAGAGCAGAGCGUUCUCGAGCGCAUUGCUCUCGUUGC ....(-(((((((((((......)))))).......(((.((..(((.((..((.....)).))..)))..)).)))))))))((((((((.(((((....))))).))))))))..... ( -45.50) >DroSim_CAF1 946 120 + 1 UAUGUUUGGCUAUAUGCUUGUGUGCAUGUAUCUGGAUCUGUUUGCCCACGGACAUGUGUAGACAGAGGGAGAGGAGAGGCCGAGAGAGCAGAGCGUUCUCGAGCGCAUUGCUCUCGUUGC .....((((((((((((......)))))).(((...(((((((((.((......)).)))))))))...))).....))))))((((((((.(((((....))))).))))))))..... ( -47.60) >consensus UAUGUUUGGCUAUAUGCUUGUGUGCAUGUAUCUGGAUCUGUUUGCCCACGGACAUGUGUAGACGGAGGGAGAGGAGAGGCCGAGAGAGCAGAGCGUUCUCGAGCGCAUUGCUCUCGUUGC .....((((((((((((......)))))).(((...(((.(((((.((((....))))..).)))).)))..)))..))))))((((((((.(((((....))))).))))))))..... (-43.57 = -43.47 + -0.11)


| Location | 7,605,529 – 7,605,649 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -30.33 |
| Energy contribution | -30.67 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
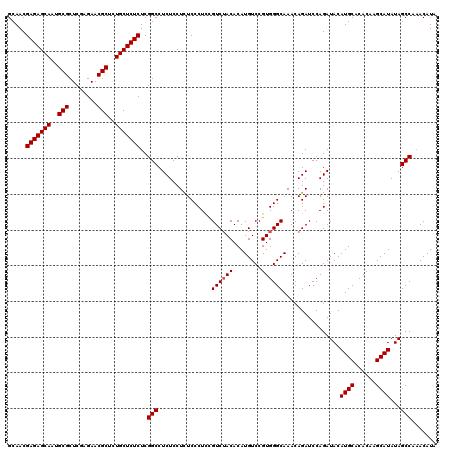
>3R_DroMel_CAF1 7605529 120 - 27905053 GCAACGAGAGCAAUGCGCUCGAGAACGCUCUGCUCUCUCGGCCUCUCCUUUCCCUUCGUCUACACAUGUCCGUGGGCAAACAGAUCCAGAUACAUGCACACAAGCAUAUAGCCAAACAUA .....(((((((..(((........)))..)))))))..(((...............((((((........))))))................((((......))))...)))....... ( -31.80) >DroSec_CAF1 945 119 - 1 GCAACGAGAGCAAUGCGCUCGAGAACGCUCUGCUCUCUCGGCCUCUCCUCUCCCUCCGUCAACACAUGUCCGUGGGCAAACAGAUCCAGAUACAUGCACACAAGCAUAUAGCCA-ACAUA .....(((((((..(((........)))..)))))))..(((.(((.....(((.(.(.((.....)).).).))).....))).........((((......))))...))).-..... ( -30.10) >DroSim_CAF1 946 120 - 1 GCAACGAGAGCAAUGCGCUCGAGAACGCUCUGCUCUCUCGGCCUCUCCUCUCCCUCUGUCUACACAUGUCCGUGGGCAAACAGAUCCAGAUACAUGCACACAAGCAUAUAGCCAAACAUA .....(((((((..(((........)))..)))))))..(((.(((..(((.....(((((((........)))))))...)))...)))...((((......))))...)))....... ( -32.70) >consensus GCAACGAGAGCAAUGCGCUCGAGAACGCUCUGCUCUCUCGGCCUCUCCUCUCCCUCCGUCUACACAUGUCCGUGGGCAAACAGAUCCAGAUACAUGCACACAAGCAUAUAGCCAAACAUA .....(((((((..(((........)))..)))))))..(((...............((((((........))))))................((((......))))...)))....... (-30.33 = -30.67 + 0.33)


| Location | 7,605,569 – 7,605,689 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -43.40 |
| Consensus MFE | -41.52 |
| Energy contribution | -42.30 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
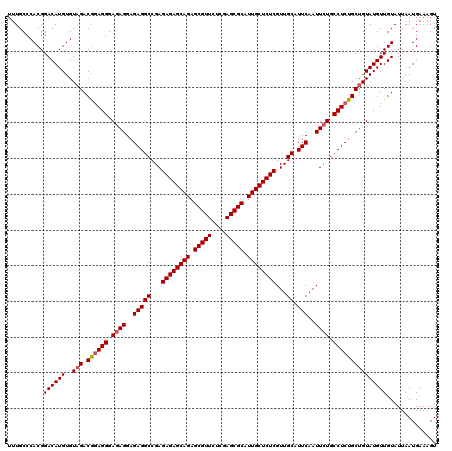
>3R_DroMel_CAF1 7605569 120 + 27905053 UUUGCCCACGGACAUGUGUAGACGAAGGGAAAGGAGAGGCCGAGAGAGCAGAGCGUUCUCGAGCGCAUUGCUCUCGUUGCAUUCAAUUCUGCCUCUGCUGUAUGUUGUAUUAAUGAAAGU ....(((.((.((....))...))..)))..((.((((((.((((((((((.(((((....))))).))))))))((((....)))))).)))))).))..................... ( -39.60) >DroSec_CAF1 984 120 + 1 UUUGCCCACGGACAUGUGUUGACGGAGGGAGAGGAGAGGCCGAGAGAGCAGAGCGUUCUCGAGCGCAUUGCUCUCGUUGCAUUCAAUUCUGCCUCUGCUGUAUGUUGUAUUAAUGAAAGU ..........((((((....(.((((((.((((..(((((...((((((((.(((((....))))).))))))))...)).)))..)))).)))))))..)))))).............. ( -43.80) >DroSim_CAF1 986 120 + 1 UUUGCCCACGGACAUGUGUAGACAGAGGGAGAGGAGAGGCCGAGAGAGCAGAGCGUUCUCGAGCGCAUUGCUCUCGUUGCAUUCAAUUCUGCCUCUGCUGUAUGUUGUAUUAAUGAAAGU ..........((((((..(((.((((((.((((..(((((...((((((((.(((((....))))).))))))))...)).)))..)))).))))))))))))))).............. ( -46.80) >consensus UUUGCCCACGGACAUGUGUAGACGGAGGGAGAGGAGAGGCCGAGAGAGCAGAGCGUUCUCGAGCGCAUUGCUCUCGUUGCAUUCAAUUCUGCCUCUGCUGUAUGUUGUAUUAAUGAAAGU ..........((((((..(((.((((((.((((..(((((...((((((((.(((((....))))).))))))))...)).)))..)))).))))))))))))))).............. (-41.52 = -42.30 + 0.78)


| Location | 7,605,569 – 7,605,689 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -33.03 |
| Energy contribution | -33.37 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
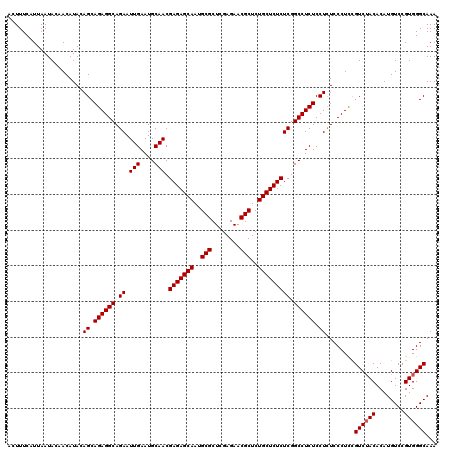
>3R_DroMel_CAF1 7605569 120 - 27905053 ACUUUCAUUAAUACAACAUACAGCAGAGGCAGAAUUGAAUGCAACGAGAGCAAUGCGCUCGAGAACGCUCUGCUCUCUCGGCCUCUCCUUUCCCUUCGUCUACACAUGUCCGUGGGCAAA .....................((.((((((.((.(((....))).(((((((..(((........)))..))))))))).)))))).))........((((((........))))))... ( -34.50) >DroSec_CAF1 984 120 - 1 ACUUUCAUUAAUACAACAUACAGCAGAGGCAGAAUUGAAUGCAACGAGAGCAAUGCGCUCGAGAACGCUCUGCUCUCUCGGCCUCUCCUCUCCCUCCGUCAACACAUGUCCGUGGGCAAA ......................((.((((.(((...((..((...(((((((..(((........)))..)))))))...)).))...))).)))).)).......((((....)))).. ( -31.90) >DroSim_CAF1 986 120 - 1 ACUUUCAUUAAUACAACAUACAGCAGAGGCAGAAUUGAAUGCAACGAGAGCAAUGCGCUCGAGAACGCUCUGCUCUCUCGGCCUCUCCUCUCCCUCUGUCUACACAUGUCCGUGGGCAAA ......................(((((((.(((...((..((...(((((((..(((........)))..)))))))...)).))...))).))))))).......((((....)))).. ( -36.50) >consensus ACUUUCAUUAAUACAACAUACAGCAGAGGCAGAAUUGAAUGCAACGAGAGCAAUGCGCUCGAGAACGCUCUGCUCUCUCGGCCUCUCCUCUCCCUCCGUCUACACAUGUCCGUGGGCAAA .....................((.((((((.((.(((....))).(((((((..(((........)))..))))))))).)))))).))........((((((........))))))... (-33.03 = -33.37 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:53 2006