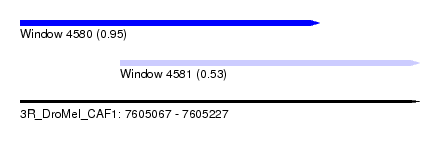
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,605,067 – 7,605,227 |
| Length | 160 |
| Max. P | 0.951222 |

| Location | 7,605,067 – 7,605,187 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.12 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -20.62 |
| Energy contribution | -20.40 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7605067 120 + 27905053 ACAGCAGAAAGGGCCAUUAAGAUUGCAUUUAUGUAUUUUAAUGGUAGGGAAUAAAUUUAUUUGCAUAACAAAAUUCUGGAUCUUGAUCACUUUGGAUGCCCUAAACUACUAAAACUACUA ....(((((...(((((((((((.(((....)))))))))))))).(.(((((....))))).).........)))))(((....)))..(((((.....)))))............... ( -21.10) >DroSec_CAF1 487 100 + 1 ACAGCAGAAAAGGCCAUUAAGAUUGCAUUUAUGUAUUUUAAUGGUAGGGAAUAAAUUUAUUUGCAUAACAAAAUUCA---------CGACUUUGGAUGCCCUUAACUAU----------- ...(((..(((((((((((((((.(((....)))))))))))))).(.(((((....))))).).............---------...))))...)))..........----------- ( -20.50) >DroSim_CAF1 487 100 + 1 ACAGCAGAAAAGGCCAUUAAGAUUGCAUUUAUGUAUUUUAAUGGUAGGGAAUAAAUUUAUUUGCAUAACAAAAUUCA---------CGACUUUGGAUGCCCUAAGCUAU----------- ..(((.......(((((((((((.(((....))))))))))))))((((..........((((.....))))(((((---------......))))).))))..)))..----------- ( -22.60) >consensus ACAGCAGAAAAGGCCAUUAAGAUUGCAUUUAUGUAUUUUAAUGGUAGGGAAUAAAUUUAUUUGCAUAACAAAAUUCA_________CGACUUUGGAUGCCCUAAACUAU___________ ...(((..(((((((((((((((.(((....)))))))))))))).(.(((((....))))).).........................))))...)))..................... (-20.62 = -20.40 + -0.22)


| Location | 7,605,107 – 7,605,227 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -19.73 |
| Consensus MFE | -18.27 |
| Energy contribution | -17.39 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.13 |
| Mean z-score | -0.29 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7605107 120 + 27905053 AUGGUAGGGAAUAAAUUUAUUUGCAUAACAAAAUUCUGGAUCUUGAUCACUUUGGAUGCCCUAAACUACUAAAACUACUAUUGUAGUUCUUUGGAGGGUAAAAGGAUAACCUGCUUUUAA ..((((((...........((((.....))))(((((.(((....)))........((((((...(((....((((((....))))))...)))))))))..)))))..))))))..... ( -24.40) >DroSec_CAF1 527 100 + 1 AUGGUAGGGAAUAAAUUUAUUUGCAUAACAAAAUUCA---------CGACUUUGGAUGCCCUUAACUAU-----------UUGUAGUUCUUUGGAGGGUAAAAGGAUAUCCUAUUUUUAA ..(((((((..........((((.....)))).....---------...((((...((((((((((((.-----------...))))).....))))))).))))...)))))))..... ( -18.10) >DroSim_CAF1 527 100 + 1 AUGGUAGGGAAUAAAUUUAUUUGCAUAACAAAAUUCA---------CGACUUUGGAUGCCCUAAGCUAU-----------UUGUAGUUCUUUGGAGGGUAAAAGGAUAACCUAUUUUCAA ..((((((((((....((((....))))....)))).---------...((((...((((((.(((((.-----------...)))))......)))))).))))....))))))..... ( -16.70) >consensus AUGGUAGGGAAUAAAUUUAUUUGCAUAACAAAAUUCA_________CGACUUUGGAUGCCCUAAACUAU___________UUGUAGUUCUUUGGAGGGUAAAAGGAUAACCUAUUUUUAA ..((((((((((....((((....))))....)))).............((((...((((((.((((((.............))))))......)))))).))))....))))))..... (-18.27 = -17.39 + -0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:48 2006