
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,579,945 – 7,580,078 |
| Length | 133 |
| Max. P | 0.998865 |

| Location | 7,579,945 – 7,580,042 |
|---|---|
| Length | 97 |
| Sequences | 6 |
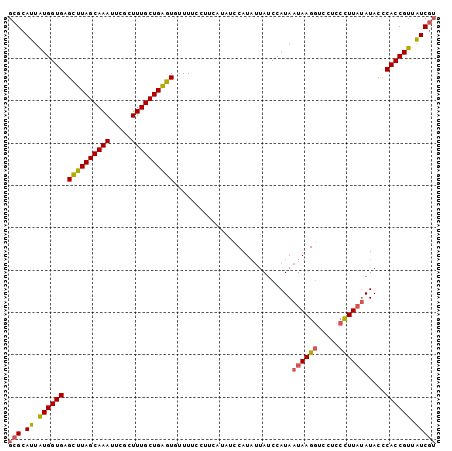
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 73.69 |
| Mean single sequence MFE | -22.59 |
| Consensus MFE | -19.07 |
| Energy contribution | -18.52 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.78 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7579945 97 + 27905053 AUUAUCUUAUUCUCCUCCGUCUGCGCAUCGUUGCGCAUUAU------GGUGAGCUUAGCAAAUUCGCUUUGCUGAGUGUUUUUCUUCAUAUCCAUAUUAUCCA ..............(.((((.((((((....))))))..))------)).).((((((((((.....)))))))))).......................... ( -24.50) >DroVir_CAF1 5909 88 + 1 AACAUUUC------GAUCUCCUGCGCAUUGUUGCGCAUUGUUGUUGUUGUUGUUGUAGCAAAUUCGCUUUGCUUCGAUUUU---------UCCAUAUUAUCAA ((((...(------(((..(.((((((....))))))..)..)))).))))((((.((((((.....)))))).))))...---------............. ( -19.50) >DroSim_CAF1 1269 97 + 1 AUUAUUUCAUUCUCCUCCUUCUGCGCAUCGUUGCGCAUUAU------GGUGAGCUUAGCAAAUUCGCUUUGCUGAGUGUUUUUCUUCAUAUCCAUAUUAUCCA ..........((.((......((((((....))))))....------)).))((((((((((.....)))))))))).......................... ( -23.40) >DroEre_CAF1 1843 97 + 1 AUUAUUCCAUUCUCCUCCUUCUGCGCAUCUUUGCGCAUUAU------GGUGAGCUUAGCAAAUUCGCUUUGCUGAGUUUUUUCCUCCAUAUCCAUUAUAUCCA .....................((((((....)))))).(((------((.((((((((((((.....))))))))))))......)))))............. ( -26.10) >DroYak_CAF1 1262 97 + 1 AUUAUCUCAUUCUCCUCCUUCUGCGCAUCUUUGCGCAUUAU------GGUGAGCUUAGCAAAUUCGCUUUGCUGAGUUUUUUCCUCCAAAUCCAUAUUAUCCA .....................((((((....)))))).(((------((.((((((((((((.....))))))))))))............)))))....... ( -25.02) >DroMoj_CAF1 5408 85 + 1 AACACGGU------UUUCUCCUGCGCAUCGUUGCGCAUUGUUGUU---GCUUUUGUAGCAAAUUCACUUUGCUUUAUUUUU---------UCCAUAUUAUCAA .....((.------.......((((((....))))))...(((((---((....)))))))....................---------.)).......... ( -17.00) >consensus AUUAUUUCAUUCUCCUCCUUCUGCGCAUCGUUGCGCAUUAU______GGUGAGCUUAGCAAAUUCGCUUUGCUGAGUUUUUU_CU_CAUAUCCAUAUUAUCCA .....................((((((....))))))...............((((((((((.....)))))))))).......................... (-19.07 = -18.52 + -0.55)
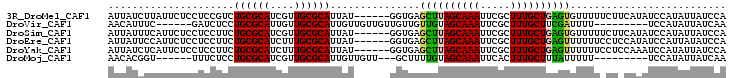
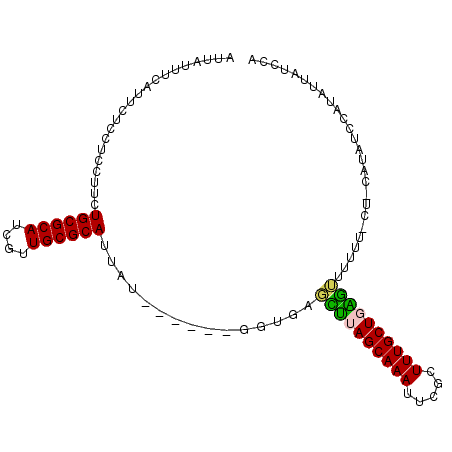

| Location | 7,579,945 – 7,580,042 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 73.69 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -14.33 |
| Energy contribution | -14.55 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7579945 97 - 27905053 UGGAUAAUAUGGAUAUGAAGAAAAACACUCAGCAAAGCGAAUUUGCUAAGCUCACC------AUAAUGCGCAACGAUGCGCAGACGGAGGAGAAUAAGAUAAU ...........................((.((((((.....)))))).))(((..(------....((((((....))))))....)..)))........... ( -20.30) >DroVir_CAF1 5909 88 - 1 UUGAUAAUAUGGA---------AAAAUCGAAGCAAAGCGAAUUUGCUACAACAACAACAACAACAAUGCGCAACAAUGCGCAGGAGAUC------GAAAUGUU .............---------....((((((((((.....))))))...................((((((....)))))).....))------))...... ( -17.90) >DroSim_CAF1 1269 97 - 1 UGGAUAAUAUGGAUAUGAAGAAAAACACUCAGCAAAGCGAAUUUGCUAAGCUCACC------AUAAUGCGCAACGAUGCGCAGAAGGAGGAGAAUGAAAUAAU ...........................((.((((((.....)))))).))(((..(------....((((((....))))))....)..)))........... ( -20.30) >DroEre_CAF1 1843 97 - 1 UGGAUAUAAUGGAUAUGGAGGAAAAAACUCAGCAAAGCGAAUUUGCUAAGCUCACC------AUAAUGCGCAAAGAUGCGCAGAAGGAGGAGAAUGGAAUAAU .................(((.......)))((((((.....))))))...(((..(------....((((((....))))))....)..)))........... ( -22.20) >DroYak_CAF1 1262 97 - 1 UGGAUAAUAUGGAUUUGGAGGAAAAAACUCAGCAAAGCGAAUUUGCUAAGCUCACC------AUAAUGCGCAAAGAUGCGCAGAAGGAGGAGAAUGAGAUAAU ...........................(((((((((.....))))))...(((..(------....((((((....))))))....)..)))...)))..... ( -22.70) >DroMoj_CAF1 5408 85 - 1 UUGAUAAUAUGGA---------AAAAAUAAAGCAAAGUGAAUUUGCUACAAAAGC---AACAACAAUGCGCAACGAUGCGCAGGAGAAA------ACCGUGUU .....(((((((.---------........((((((.....))))))........---........((((((....)))))).......------.))))))) ( -20.20) >consensus UGGAUAAUAUGGAUAUG_AG_AAAAAACUCAGCAAAGCGAAUUUGCUAAGCUCACC______AUAAUGCGCAACGAUGCGCAGAAGGAGGAGAAUGAAAUAAU ...........................(((((((((.....))))))...................((((((....))))))....))).............. (-14.33 = -14.55 + 0.22)

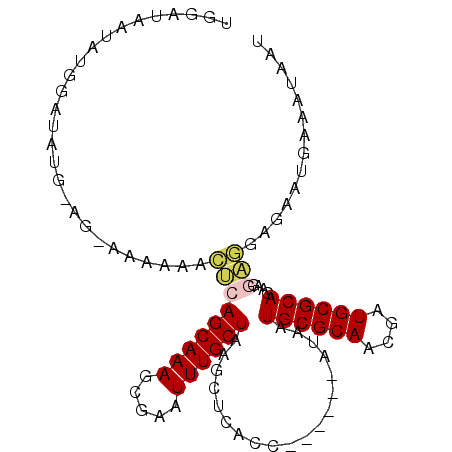

| Location | 7,579,977 – 7,580,078 |
|---|---|
| Length | 101 |
| Sequences | 6 |
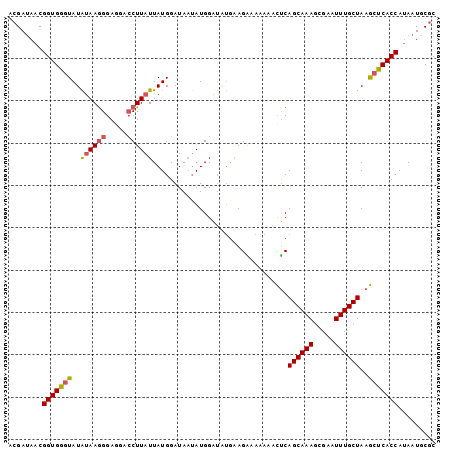
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 90.10 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -22.08 |
| Energy contribution | -22.08 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
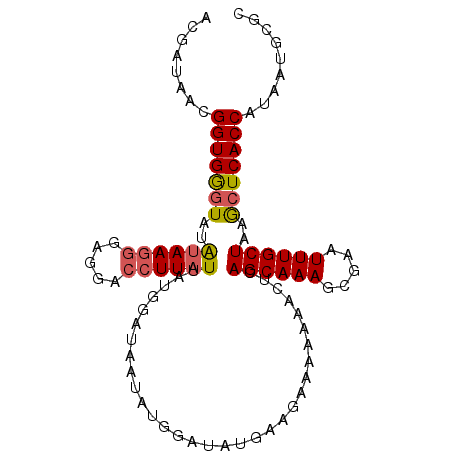
>3R_DroMel_CAF1 7579977 101 + 27905053 GCGCAUUAUGGUGAGCUUAGCAAAUUCGCUUUGCUGAGUGUUUUUCUUCAUAUCCAUAUUAUCCAUAAUAAGGUCCUCCCUUAUAUACCCACCGUUAUCGU (((.((.((((((.((((((((((.....))))))))))............................((((((.....)))))).....)))))).))))) ( -25.60) >DroSec_CAF1 1294 101 + 1 GCGCAUUAUGGUGAGCUUAGCAAAUUCGCUUUGCUGAGUGUUUUUCUUCAUAUCCAUAUUAUCCAUAAUAAGGUCCUCCCUUAUAUAUCCACCGUUAUCGU (((.((.((((((.((((((((((.....))))))))))............................((((((.....)))))).....)))))).))))) ( -25.60) >DroSim_CAF1 1301 101 + 1 GCGCAUUAUGGUGAGCUUAGCAAAUUCGCUUUGCUGAGUGUUUUUCUUCAUAUCCAUAUUAUCCAUAAUAAGGUCCUCCCUUAUAUACCCACCGUUAUCGU (((.((.((((((.((((((((((.....))))))))))............................((((((.....)))))).....)))))).))))) ( -25.60) >DroEre_CAF1 1875 101 + 1 GCGCAUUAUGGUGAGCUUAGCAAAUUCGCUUUGCUGAGUUUUUUCCUCCAUAUCCAUUAUAUCCAUAAUAAGGUCAUCCCUUAUAUACCCACCGUUAUCGU (((.((.(((((((((((((((((.....)))))))))))...........................((((((.....)))))).....)))))).))))) ( -25.70) >DroYak_CAF1 1294 101 + 1 GCGCAUUAUGGUGAGCUUAGCAAAUUCGCUUUGCUGAGUUUUUUCCUCCAAAUCCAUAUUAUCCAUAAUAAGGUCCUCCCUUAUAUACCCACCGUUAUCGU (((.((.(((((((((((((((((.....)))))))))))...........................((((((.....)))))).....)))))).))))) ( -25.70) >DroAna_CAF1 1400 101 + 1 GCGCAUUGUGGUGAGUUUAGCAAAUUCACUUUGCUGGAUUGUUUCCUUUUCUCCCAUAUUAUCCAUAAUAAGCUGCUCCUUUCCAUACUCACCGUUGUCUA ..(((...((((((((..((((((.....))))))(((.....)))........................................)))))))).)))... ( -18.90) >consensus GCGCAUUAUGGUGAGCUUAGCAAAUUCGCUUUGCUGAGUGUUUUCCUUCAUAUCCAUAUUAUCCAUAAUAAGGUCCUCCCUUAUAUACCCACCGUUAUCGU (((.((.((((((.((((((((((.....))))))))))............................((((((.....)))))).....)))))).))))) (-22.08 = -22.08 + 0.00)

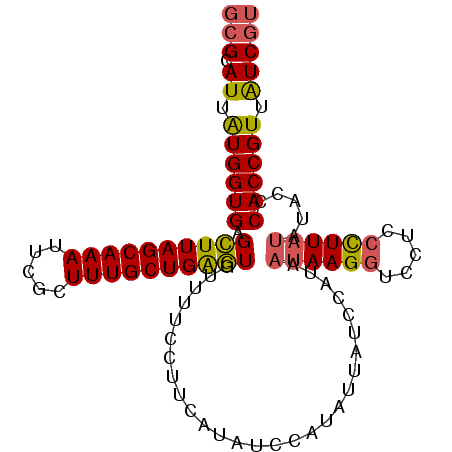
| Location | 7,579,977 – 7,580,078 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 90.10 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -19.68 |
| Energy contribution | -19.93 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.595695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7579977 101 - 27905053 ACGAUAACGGUGGGUAUAUAAGGGAGGACCUUAUUAUGGAUAAUAUGGAUAUGAAGAAAAACACUCAGCAAAGCGAAUUUGCUAAGCUCACCAUAAUGCGC ........(((((((..((((((.....))))))................................((((((.....))))))..)))))))......... ( -22.30) >DroSec_CAF1 1294 101 - 1 ACGAUAACGGUGGAUAUAUAAGGGAGGACCUUAUUAUGGAUAAUAUGGAUAUGAAGAAAAACACUCAGCAAAGCGAAUUUGCUAAGCUCACCAUAAUGCGC ........(((((.(((((((((.....))))).)))).........................((.((((((.....)))))).)).)))))......... ( -18.80) >DroSim_CAF1 1301 101 - 1 ACGAUAACGGUGGGUAUAUAAGGGAGGACCUUAUUAUGGAUAAUAUGGAUAUGAAGAAAAACACUCAGCAAAGCGAAUUUGCUAAGCUCACCAUAAUGCGC ........(((((((..((((((.....))))))................................((((((.....))))))..)))))))......... ( -22.30) >DroEre_CAF1 1875 101 - 1 ACGAUAACGGUGGGUAUAUAAGGGAUGACCUUAUUAUGGAUAUAAUGGAUAUGGAGGAAAAAACUCAGCAAAGCGAAUUUGCUAAGCUCACCAUAAUGCGC ........(((((((..((((((.....))))))...................(((.......)))((((((.....))))))..)))))))......... ( -24.60) >DroYak_CAF1 1294 101 - 1 ACGAUAACGGUGGGUAUAUAAGGGAGGACCUUAUUAUGGAUAAUAUGGAUUUGGAGGAAAAAACUCAGCAAAGCGAAUUUGCUAAGCUCACCAUAAUGCGC ........(((((((..((((((.....))))))...................(((.......)))((((((.....))))))..)))))))......... ( -24.60) >DroAna_CAF1 1400 101 - 1 UAGACAACGGUGAGUAUGGAAAGGAGCAGCUUAUUAUGGAUAAUAUGGGAGAAAAGGAAACAAUCCAGCAAAGUGAAUUUGCUAAACUCACCACAAUGCGC ........(((((((.......(((....(((..(((.....)))...)))....(....)..)))((((((.....))))))..)))))))......... ( -23.90) >consensus ACGAUAACGGUGGGUAUAUAAGGGAGGACCUUAUUAUGGAUAAUAUGGAUAUGAAGAAAAAAACUCAGCAAAGCGAAUUUGCUAAGCUCACCAUAAUGCGC ........(((((((..((((((.....))))))................................((((((.....))))))..)))))))......... (-19.68 = -19.93 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:43 2006