
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,575,531 – 7,575,744 |
| Length | 213 |
| Max. P | 0.907319 |

| Location | 7,575,531 – 7,575,647 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.52 |
| Mean single sequence MFE | -44.90 |
| Consensus MFE | -36.96 |
| Energy contribution | -37.20 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
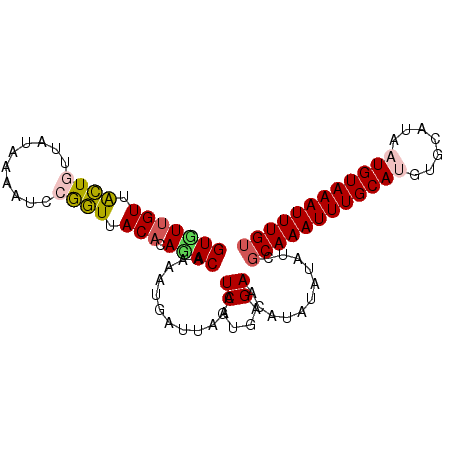
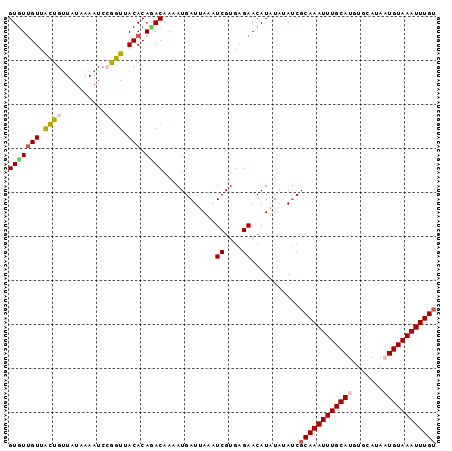
>3R_DroMel_CAF1 7575531 116 + 27905053 GAGUGGGUGGUGGGGGUUGGC----AGGCCAACAGCUGCUAUUGGGGAACACAAGGGAGUUUAAAUGCAAUUCUUUUCGCGGGGGCGGGGGACUAUUCUCAUUCGCAGCUGCUGUCCGGA ...((((..((....(((((.----...)))))((((((...((((((.......((((((.......))))))(..(((....)))..).....))))))...))))))))..)))).. ( -38.60) >DroSec_CAF1 7547 116 + 1 GAGUGGGUGGCGGGGGUUGGC----AGGCCAACAGCUGCUAUUGGGGAACAAAAGGGAGCUUAAAUGCAAUUCUUUUCGCGGGGGCGGGGGACUAUUCUCAUUCGCAGCUGCUGCCCGGA ...((((..((....(((((.----...)))))((((((...((((((..........((((...(((..........))).))))((....)).))))))...))))))))..)))).. ( -44.70) >DroSim_CAF1 7984 116 + 1 GAGUGGGUGGCGGGGGUUGGC----AGGCCAACAGCUGCUAUUGGGGAACAAAAGGGAGCUUAAAUGCAAUUCUUUUCGCGGGGGCGGGGGACUAUUCUCAUUCGCAGCUGCUGCCCGGA ...((((..((....(((((.----...)))))((((((...((((((..........((((...(((..........))).))))((....)).))))))...))))))))..)))).. ( -44.70) >DroEre_CAF1 7982 111 + 1 U---------CGGGGGUUGGCCAACUGGCCAACAGCUGCUAUUGGGGAACAAAAGGGAGCUGAAAUGCAAUUCUCUUCGUGGGGGCGGGGUACUGUUCUCAUUCGCAGCAGCUGCCCUGA (---------((((((((((((....)))))))(((((((....(.((....(((((((.((.....)).))))))).((((((((((....)))))))))))).)))))))).)))))) ( -51.00) >DroYak_CAF1 7645 116 + 1 GAGUGGGUGGUGGGGGUUGGC----AGGCCAACAGCUGCUAUUGGGGAACAAAAGGGAGCUCAAAUGCAAUUCUUUUUGAGAGGGCGGGGUACUCUUCUCAUAAGCAGCUGCUGCCCUGA ....(((..((....(((((.----...)))))(((((((..((((((.(((((((((((......))..))))))))).(((..(...)..)))))))))..)))))))))..)))... ( -45.50) >consensus GAGUGGGUGGCGGGGGUUGGC____AGGCCAACAGCUGCUAUUGGGGAACAAAAGGGAGCUUAAAUGCAAUUCUUUUCGCGGGGGCGGGGGACUAUUCUCAUUCGCAGCUGCUGCCCGGA ....(((..((....((((((......))))))((((((...((((((..........((......))......(..(((....)))..).....))))))...))))))))..)))... (-36.96 = -37.20 + 0.24)



| Location | 7,575,531 – 7,575,647 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.52 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -25.42 |
| Energy contribution | -26.54 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7575531 116 - 27905053 UCCGGACAGCAGCUGCGAAUGAGAAUAGUCCCCCGCCCCCGCGAAAAGAAUUGCAUUUAAACUCCCUUGUGUUCCCCAAUAGCAGCUGUUGGCCU----GCCAACCCCCACCACCCACUC ...((.((((((((((...((.((((..............((((......))))......((......))))))..))...)))))))))).)).----..................... ( -26.40) >DroSec_CAF1 7547 116 - 1 UCCGGGCAGCAGCUGCGAAUGAGAAUAGUCCCCCGCCCCCGCGAAAAGAAUUGCAUUUAAGCUCCCUUUUGUUCCCCAAUAGCAGCUGUUGGCCU----GCCAACCCCCGCCACCCACUC ...(((..((((((((...((.((((.......(((....)))(((((....((......))...)))))))))..))...))))))(((((...----.)))))....))..))).... ( -32.80) >DroSim_CAF1 7984 116 - 1 UCCGGGCAGCAGCUGCGAAUGAGAAUAGUCCCCCGCCCCCGCGAAAAGAAUUGCAUUUAAGCUCCCUUUUGUUCCCCAAUAGCAGCUGUUGGCCU----GCCAACCCCCGCCACCCACUC ...(((..((((((((...((.((((.......(((....)))(((((....((......))...)))))))))..))...))))))(((((...----.)))))....))..))).... ( -32.80) >DroEre_CAF1 7982 111 - 1 UCAGGGCAGCUGCUGCGAAUGAGAACAGUACCCCGCCCCCACGAAGAGAAUUGCAUUUCAGCUCCCUUUUGUUCCCCAAUAGCAGCUGUUGGCCAGUUGGCCAACCCCCG---------A ...(((.((((((((.....(.((((.......((......))(((((....((......))...))))))))))....))))))))(((((((....))))))).))).---------. ( -34.50) >DroYak_CAF1 7645 116 - 1 UCAGGGCAGCAGCUGCUUAUGAGAAGAGUACCCCGCCCUCUCAAAAAGAAUUGCAUUUGAGCUCCCUUUUGUUCCCCAAUAGCAGCUGUUGGCCU----GCCAACCCCCACCACCCACUC .((((.(((((((((((..(((((.(.((.....))).)))))(((((....((......))...)))))..........))))))))))).)))----).................... ( -35.80) >consensus UCCGGGCAGCAGCUGCGAAUGAGAAUAGUCCCCCGCCCCCGCGAAAAGAAUUGCAUUUAAGCUCCCUUUUGUUCCCCAAUAGCAGCUGUUGGCCU____GCCAACCCCCGCCACCCACUC ...((.((((((((((...((.((((.......(((....)))(((((....((......))...)))))))))..))...)))))))))).)).......................... (-25.42 = -26.54 + 1.12)



| Location | 7,575,647 – 7,575,744 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 89.38 |
| Mean single sequence MFE | -20.16 |
| Consensus MFE | -15.96 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7575647 97 - 27905053 GUGUUGUUACUGCUAUAAAAUCCAGUAACACAUACAAAAUGAUUAAAUCGUGAGAACAUAUAUAUCGCAAAUUUGCAUGUGCAUAAUGUAAAUUUGU ((((((((((((..........)))))))).))))............((....))...........((((((((((((.......)))))))))))) ( -24.40) >DroSec_CAF1 7663 97 - 1 GUGUUGUUACUGUUAUAAAAUCCGGUUACACAUACAAAAUGAUUAAAUCGUGAGAACAUAUAUAUCGCAAAUUUGCAUGUGCAUAAUGUAAAUUUGU (((((((.((((..........)))).))).))))............((....))...........((((((((((((.......)))))))))))) ( -20.00) >DroSim_CAF1 8100 97 - 1 GUGUUGUUACUGUUAUAAAAUCCGGUUACACACACAAAAUGAUUAAAUCGUGAGAACAUAUAUAUCGCAAAUUUGCAUGUGCAUAAUGUAAAUUUGU (((((((.((((..........)))).))).))))............((....))...........((((((((((((.......)))))))))))) ( -21.90) >DroEre_CAF1 8093 97 - 1 GUUUUGUUAAUGUUAUAAAAUCGGUUUACACAGACAAAAUGAUAACGUCGUGAGAACAUUUAAAUCGCAAAUUUGCAUCUGCAAAUUGUAAAUUUGG (((((((.......)))))))..((((.(((.(((...........)))))).))))..........((((((((((.........)))))))))). ( -17.00) >DroYak_CAF1 7761 97 - 1 GUGUAGUUGCUGUUAUAAAAUCGGGUUACACAGACAAAAUGAUAAGCUCGUGAGAACAUUUAAAACGCAAAUUUGCAUGUGCAGAUUGUAAAUUUGU ((((((...(((.........))).))))))..............(.((....)).).........(((((((((((.........))))))))))) ( -17.50) >consensus GUGUUGUUACUGUUAUAAAAUCCGGUUACACAGACAAAAUGAUUAAAUCGUGAGAACAUAUAUAUCGCAAAUUUGCAUGUGCAUAAUGUAAAUUUGU (((((((.((((..........)))).))).))))............((....))...........((((((((((((.......)))))))))))) (-15.96 = -16.40 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:38 2006