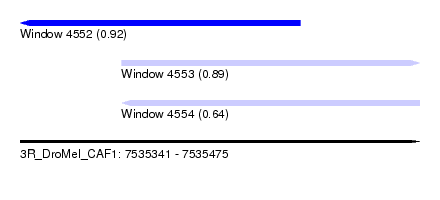
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,535,341 – 7,535,475 |
| Length | 134 |
| Max. P | 0.917216 |

| Location | 7,535,341 – 7,535,435 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.71 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -12.70 |
| Energy contribution | -13.43 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
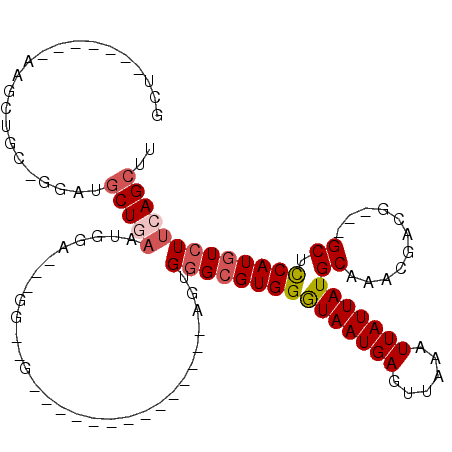
>3R_DroMel_CAF1 7535341 94 - 27905053 GCUGGAGCUGAAGCUGCAGGAUGCUGAAUGGA---AG--G--------------AGUGGGCGUGGGUAAUGAGUUAAAUUAUUAUGCAAACGACG---GCUUCAUGUCUUCAGCUU ((((((((((((((((..(.(((((.(.....---..--.--------------..).)))))(.(((((((......))))))).)...)..))---)))))).).))))))).. ( -32.10) >DroSec_CAF1 30728 94 - 1 GCUGGAGCUGAAGCUGCUGGAUGCUGAAUGGA---GG--G--------------AGUGGGCGUGGGUAAUGAGUUAAAUUAUUAUGCAAACGACG---GCUCCAUGUCUUCAGCUU .(..((((....))).)..)..((((((.((.---.(--(--------------(((.(.((((.(((((((......))))))).)..))).).---)))))...)))))))).. ( -32.70) >DroSim_CAF1 31611 79 - 1 GCU---------------AGAUGCUGAAUGGA---GG--G--------------AGUGGGCGUGGGUAAUGAGUUAAAUUAUUAUGCAAACGACG---GCUCCAUGUCUUCAGCUU ...---------------....((((((.((.---.(--(--------------(((.(.((((.(((((((......))))))).)..))).).---)))))...)))))))).. ( -27.00) >DroEre_CAF1 32585 87 - 1 GCU------AAAGCUGCA-GAAGCUGAAUAGA---GG--G--------------AGUGAGCGUGGGUAAUGAGUUAAAUUAUUAUGCAAACGACG---GCUCCAUGUCUUCAGCUU ((.------......)).-.((((((((....---.(--(--------------(((...((((.(((((((......))))))).)..)))...---))))).....)))))))) ( -26.60) >DroYak_CAF1 30676 98 - 1 GCU------AAAGCUA----AAGCUAAAUAGA---GA--GAGAGAGAGAGUGGGAAUGGGCGUGGGUAAUGAGUUAAAUUAUUAUGCAAACGACG---GCUCCAUGUCUUCAGCUU (((------..(((..----..))).......---((--.(((.(..((((.(......((((((.((((.......))))))))))......).---))))..).)))))))).. ( -21.90) >DroPer_CAF1 30025 86 - 1 ---------------GGUGCAUGCUGCCUGGUGCAGGCCA--------------AGAGGGAGUGGAUAAUGAGUUAAAUUAUUAUGCAAACGACGGCGGCUCCAUGUCUUGAGA-U ---------------..........(((((...)))))((--------------(((.((((((.(((((((......))))))).)...((....)))))))...)))))...-. ( -23.90) >consensus GCU_______AAGCUGC_GGAUGCUGAAUGGA___GG__G______________AGUGGGCGUGGGUAAUGAGUUAAAUUAUUAUGCAAACGACG___GCUCCAUGUCUUCAGCUU ......................(((((..............................(((((((((((((((......)))))))((...........)).))))))))))))).. (-12.70 = -13.43 + 0.72)

| Location | 7,535,375 – 7,535,475 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.60 |
| Mean single sequence MFE | -19.56 |
| Consensus MFE | -10.71 |
| Energy contribution | -11.78 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
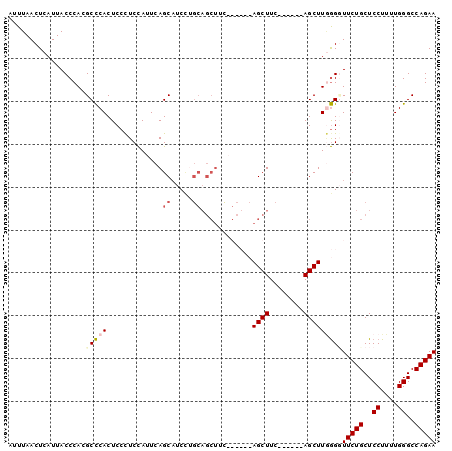
>3R_DroMel_CAF1 7535375 100 + 27905053 AUUUAACUCAUUACCCACGCCCACUCCUUCCAUUCAGCAUCCUGCAGCUUCAGCUCCAGCUUCUGCUCUAGCUUAGGGUUCUGCUCCUUUUGGGCCAGAA ..................(((((...........(((.((((((.((((..(((..........)))..)))))))))).))).......)))))..... ( -23.97) >DroSec_CAF1 30762 94 + 1 AUUUAACUCAUUACCCACGCCCACUCCCUCCAUUCAGCAUCCAGCAGCUUCAGCUCCAGCUUC------AGCUUGGGGUUCUGCUCCUUAUGGGCCAGAA ..................(((((.............((.....))(((...(((((((((...------.)).)))))))..))).....)))))..... ( -23.10) >DroSim_CAF1 31645 79 + 1 AUUUAACUCAUUACCCACGCCCACUCCCUCCAUUCAGCAUCU---------------AGCUUC------AGCUUGGGUUUCUGCUCCUUUUGGGCCAGAA ............(((((.((...............(((....---------------.)))..------.)).)))))(((((..((....))..))))) ( -14.97) >DroEre_CAF1 32619 87 + 1 AUUUAACUCAUUACCCACGCUCACUCCCUCUAUUCAGCUUC-UGCAGCUUU------AGCUUC------AGCUUCAGCUUCUGCUCCUUUUGGGCCAGAA ....................................((...-.))((((..------(((...------.)))..))))((((..((....))..)))). ( -16.20) >consensus AUUUAACUCAUUACCCACGCCCACUCCCUCCAUUCAGCAUCCUGCAGCUUC______AGCUUC______AGCUUGGGGUUCUGCUCCUUUUGGGCCAGAA ..................(((((.............((.....))............((((........)))))))))(((((..((....))..))))) (-10.71 = -11.78 + 1.06)

| Location | 7,535,375 – 7,535,475 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.60 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -15.41 |
| Energy contribution | -15.97 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
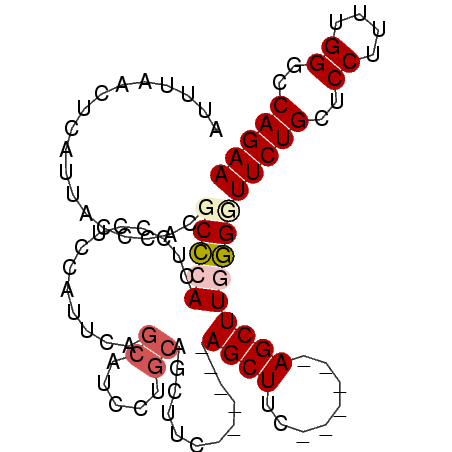
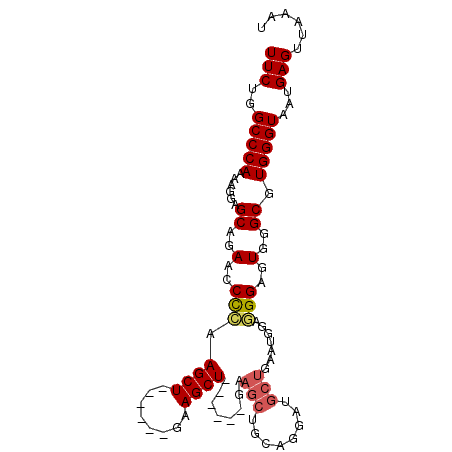
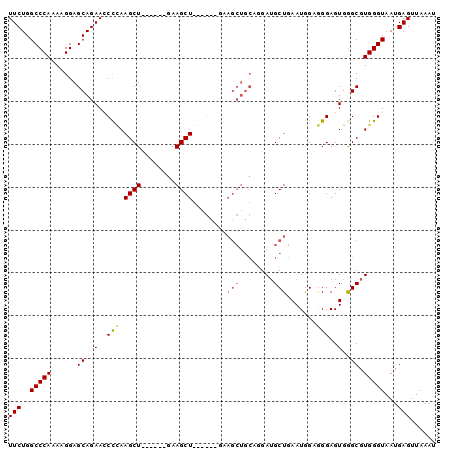
>3R_DroMel_CAF1 7535375 100 - 27905053 UUCUGGCCCAAAAGGAGCAGAACCCUAAGCUAGAGCAGAAGCUGGAGCUGAAGCUGCAGGAUGCUGAAUGGAAGGAGUGGGCGUGGGUAAUGAGUUAAAU (((..(((((......((...(((((...(((.((((....(((.(((....))).)))..))))...))).))).))..)).)))))...)))...... ( -29.20) >DroSec_CAF1 30762 94 - 1 UUCUGGCCCAUAAGGAGCAGAACCCCAAGCU------GAAGCUGGAGCUGAAGCUGCUGGAUGCUGAAUGGAGGGAGUGGGCGUGGGUAAUGAGUUAAAU (((..((((((.....((...(((((.((((------..(((....)))..))))((.....))........))).))..))))))))...)))...... ( -28.20) >DroSim_CAF1 31645 79 - 1 UUCUGGCCCAAAAGGAGCAGAAACCCAAGCU------GAAGCU---------------AGAUGCUGAAUGGAGGGAGUGGGCGUGGGUAAUGAGUUAAAU (((((..((....))..)))))(((((.(((------.(..((---------------...(.(.....).).))..).))).)))))............ ( -20.50) >DroEre_CAF1 32619 87 - 1 UUCUGGCCCAAAAGGAGCAGAAGCUGAAGCU------GAAGCU------AAAGCUGCA-GAAGCUGAAUAGAGGGAGUGAGCGUGGGUAAUGAGUUAAAU (((..(((((......((((.((((......------..))))------....)))).-...(((.(..........).))).)))))...)))...... ( -20.60) >consensus UUCUGGCCCAAAAGGAGCAGAACCCCAAGCU______GAAGCU______GAAGCUGCAGGAUGCUGAAUGGAGGGAGUGGGCGUGGGUAAUGAGUUAAAU (((..(((((......((..(..(((.((((........))))........(((........))).......)))..)..)).)))))...)))...... (-15.41 = -15.97 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:22 2006