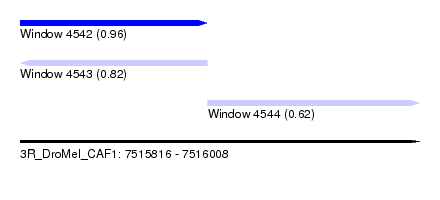
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,515,816 – 7,516,008 |
| Length | 192 |
| Max. P | 0.960135 |

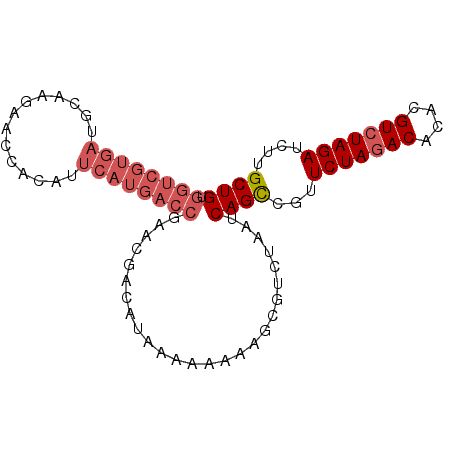
| Location | 7,515,816 – 7,515,906 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 80.37 |
| Mean single sequence MFE | -21.84 |
| Consensus MFE | -16.33 |
| Energy contribution | -18.39 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7515816 90 + 27905053 UGGUCGUGAUGCAAAAGCCACAUUCAUGACCGAACGACUUAAAAAAAAGCGUCGUAUCAGUCGUUCUAAACAAACGUCUAGAUCAUGCUG .((((((((.((....)).....))))))))................(((((.(.(((((.((((.......)))).)).))))))))). ( -22.30) >DroSec_CAF1 11467 89 + 1 UGGUCGUGAUGCAAGAACCACAUUCAUGACCGAACGAAAUAAA-AAAAGCGUCUAAUCAGCCGUUCUAGACACACGUCUAGAUCUUGCUG .((((((((..............))))))))............-.............((((...(((((((....)))))))....)))) ( -24.54) >DroSim_CAF1 11970 90 + 1 UGGUCGUGAUGCAAGAACCACAUUCAUGACCGAACGACUUAAAAAAAAGCGUCUAAUCAGCCGUUCUAGACACACGUCUAGAUCUUGCUG .((((((((..............))))))))((.((.(((......)))))))....((((...(((((((....)))))))....)))) ( -26.94) >DroEre_CAF1 11544 70 + 1 ------------------CACAUGCACGCCCGAACGACAA-GAAAAAAGCGUCUAAUCAGCCGUUCUAGACA-ACGUCUAGAUCUUGCUG ------------------.......((((...........-.......)))).....((((...(((((((.-..)))))))....)))) ( -13.57) >consensus UGGUCGUGAUGCAAGAACCACAUUCAUGACCGAACGACAUAAAAAAAAGCGUCUAAUCAGCCGUUCUAGACACACGUCUAGAUCUUGCUG .((((((((..............))))))))..........................((((...(((((((....)))))))....)))) (-16.33 = -18.39 + 2.06)

| Location | 7,515,816 – 7,515,906 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 80.37 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -18.36 |
| Energy contribution | -20.18 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7515816 90 - 27905053 CAGCAUGAUCUAGACGUUUGUUUAGAACGACUGAUACGACGCUUUUUUUUAAGUCGUUCGGUCAUGAAUGUGGCUUUUGCAUCACGACCA ...((((((((((((....)))))((((((((...................)))))))))))))))..((((((....))..)))).... ( -24.11) >DroSec_CAF1 11467 89 - 1 CAGCAAGAUCUAGACGUGUGUCUAGAACGGCUGAUUAGACGCUUUU-UUUAUUUCGUUCGGUCAUGAAUGUGGUUCUUGCAUCACGACCA ((((....(((((((....)))))))...)))).............-............((((.(((.((..(...)..))))).)))). ( -24.80) >DroSim_CAF1 11970 90 - 1 CAGCAAGAUCUAGACGUGUGUCUAGAACGGCUGAUUAGACGCUUUUUUUUAAGUCGUUCGGUCAUGAAUGUGGUUCUUGCAUCACGACCA ((((....(((((((....)))))))...))))....(((((((......))).)))).((((.(((.((..(...)..))))).)))). ( -28.60) >DroEre_CAF1 11544 70 - 1 CAGCAAGAUCUAGACGU-UGUCUAGAACGGCUGAUUAGACGCUUUUUUC-UUGUCGUUCGGGCGUGCAUGUG------------------ ..((....(((((((..-.)))))))(((.((((...((((........-.))))..)))).))))).....------------------ ( -21.50) >consensus CAGCAAGAUCUAGACGUGUGUCUAGAACGGCUGAUUAGACGCUUUUUUUUAAGUCGUUCGGUCAUGAAUGUGGUUCUUGCAUCACGACCA ..(((((((((((((....)))))))..((((((...(((............)))..))))))...........)))))).......... (-18.36 = -20.18 + 1.81)

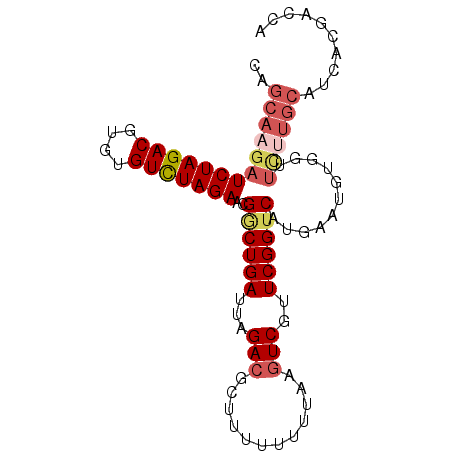
| Location | 7,515,906 – 7,516,008 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.62 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -12.53 |
| Energy contribution | -13.73 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
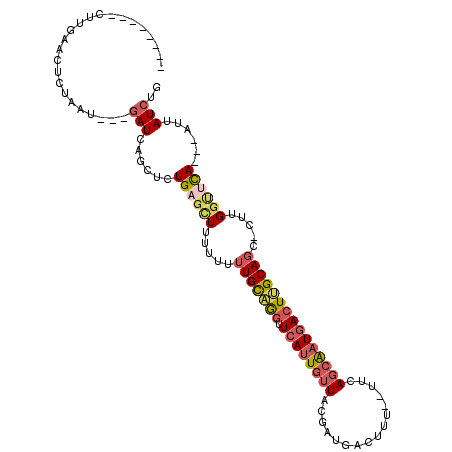
>3R_DroMel_CAF1 7515906 102 + 27905053 --------CUUGAAACCUAAUGAUGAUCAGCACUGAGCUUUUUUUUGCAGUUUCAUUGUUACAAUGGCUUU--UUCAGCAAUGACUUGCAGC-CUUGGUUCAACAAUUAUCUG --------.(((((.(((((..(((((((....)))((........))....))))..)))....((((..--.(((....))).....)))-)..))))))).......... ( -20.00) >DroSec_CAF1 11556 96 + 1 --------CUUGAACUCUAAU---GAUCAGCUCUGAGCUUUUUUUUGCAGGUUCAUUGUUACGAUGACUUU--UUUAGCAAUGACUUGCAGC-CUUGGUUCA---AUUAUCUG --------.(((((((.....---....(((.....))).....((((((((.((((((((.((......)--).)))))))))))))))).-...))))))---)....... ( -25.30) >DroSim_CAF1 12060 96 + 1 --------CUUGAACUCUAAU---GAUCAGCGCUGAGCUUUUUUUUGCAGGUUCAUUGUUACGAUGACUUU--UUGAGCAAUGACUUGCAGC-CUUGGUUCA---AUUAUCUG --------.(((((((.....---....(((.....))).....((((((((.(((((((.(((.......--)))))))))))))))))).-...))))))---)....... ( -25.70) >DroYak_CAF1 11545 96 + 1 --------ACUGAACUUCAGU---GAUCACCUCUGUGCUUUUUUUUGCAGGCUCACUGUUACGGCGACUUU--CUCAGCAAUGACUGGCAGC-CUUGACUUA---AUUAUCUG --------..........(((---((...(((....((........))))).)))))((((.(((..((..--.(((....)))..))..))-).))))...---........ ( -19.30) >DroAna_CAF1 11683 105 + 1 UUUUGAAUCUCCAAACUUGCU---AAUCAAGUUUGUAUUGCUUUGUGUUAGUUAAUUAUUACG--GACUGCAAGUAAUCGAUGACUGGCAAAAAGUGGGUUA---UUUAUCUC .....(((((.((((((((..---...))))))))....(((((.(((((((((.(.(((((.--........))))).).))))))))).)))))))))).---........ ( -24.40) >consensus ________CUUGAACUCUAAU___GAUCAGCUCUGAGCUUUUUUUUGCAGGUUCAUUGUUACGAUGACUUU__UUCAGCAAUGACUUGCAGC_CUUGGUUCA___AUUAUCUG ........................(((......((((((.....(((((((.((((((((................))))))))))))))).....))))))......))).. (-12.53 = -13.73 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:12 2006