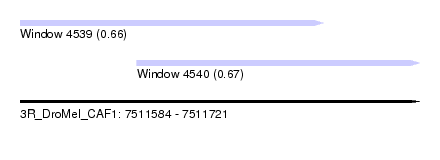
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,511,584 – 7,511,721 |
| Length | 137 |
| Max. P | 0.667477 |

| Location | 7,511,584 – 7,511,688 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.82 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -18.23 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7511584 104 + 27905053 UUUCCAUUUCCUUGGGAAACAUAUUAACGUCCGUAAAGUUGUCGGGCGUGGACUUUGGCUGCACUUUCAGUAUGUAGGAGAACGUCUUGGAAGUGUGAUCUGCA------- ...(((..(((...(....)......(((((((.........))))))))))...)))..((((((((((.((((......)))).))))))))))........------- ( -30.70) >DroSec_CAF1 7275 104 + 1 UUUCCCUUUCCUUGGGAAACAUAUUAACGUCCGUAAAGUUGUCGGGCGUGGACUGUGGCAUCACUUUCAGGAUGUAGGACAACGUCUUGGAUGUGUUAUCUGCA------- ((((((.......))))))((((...(((((((.........)))))))....))))(((.(((.((((((((((......)))))))))).))).....))).------- ( -36.10) >DroSim_CAF1 7625 104 + 1 UUUCCCUUUCCUUGGGAAACAUAUUAACGUCCGUAAAGUUGUCGGGCGUGGCCUGUGGCAUCACUUUCAGGAUGUAGGAAAACGUCUUGGAAGUGUUAUCUGCA------- ((((((.......))))))((((...(((((((.........)))))))....))))(((.((((((((((((((......)))))))))))))).....))).------- ( -40.20) >DroEre_CAF1 7341 104 + 1 UUUCCAUUUCUUUGGGAAACAUAUUAGCAUCGUUAAAGUUGUCGGGUGUGGAGCUCGGCUGUACUUUCAGGAUGUAGGAGAACGUCUUGGCAGUGUUAUCUGCA------- (((((.........(....)......(((((.(.(((((.(((((((.....)))))))...))))).).))))).)))))........((((......)))).------- ( -31.80) >DroYak_CAF1 7373 104 + 1 UUUCCAUUUCCUUGGGAAACAUAUUGACGUCGGUAAAGUUAUCGGGAGUGGACUUCGGCAGCACUUUCAGGAUGUAGGAGAAGGUCUUGGCAGUGUUAUCUGCA------- ..(((((((((...(....).(((((....)))))........))))))))).....((((((((.(((((((.(......).))))))).))))....)))).------- ( -32.20) >DroAna_CAF1 7383 111 + 1 UUUCCAUUUCUUUUGGAAACAUAUUAAAAUCUGCGAAAUUCUCCCCCGUGUCUGGGAGUUUUACUUUUAGGAUGUAGGAGAAUGUCUUUGAACUGCUGUCUGUUAAUUAAA ((((((.......))))))..........((...((.(((((((..(((.((((((((.....)))))))))))..))))))).))...)).................... ( -25.00) >consensus UUUCCAUUUCCUUGGGAAACAUAUUAACGUCCGUAAAGUUGUCGGGCGUGGACUGUGGCAGCACUUUCAGGAUGUAGGAGAACGUCUUGGAAGUGUUAUCUGCA_______ (((((.........))))).............(((.....((((((.......))))))..((((((((((((((......)))))))))))))).....)))........ (-18.23 = -18.68 + 0.45)



| Location | 7,511,624 – 7,511,721 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 73.89 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -17.07 |
| Energy contribution | -17.88 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7511624 97 + 27905053 GUCGGGCGUGGACUUUGGCUGCACUUUCAGUAUGUAGGAGAACGUCUUGGAAGUGUGAUCUGCA-------AAGGA----UUUUCUCGAUUAAGCCCACUCUCACACU ((.(((.((((.(((...(.((((((((((.((((......)))).))))))))))(((((...-------..)))----)).....)...))).)))).)))))... ( -28.00) >DroSec_CAF1 7315 97 + 1 GUCGGGCGUGGACUGUGGCAUCACUUUCAGGAUGUAGGACAACGUCUUGGAUGUGUUAUCUGCA-------UAGGA----UUCUCUUGAUUAAACGUACUCUCACACU ((((((.(.(..(((((.((.(((.((((((((((......)))))))))).))).....))))-------)))..----).).)))))).................. ( -25.00) >DroSim_CAF1 7665 97 + 1 GUCGGGCGUGGCCUGUGGCAUCACUUUCAGGAUGUAGGAAAACGUCUUGGAAGUGUUAUCUGCA-------UAGGA----UUUUCUUUAUUAAACCCACUCUCACACU ((.(((.((((((((((.((.((((((((((((((......)))))))))))))).....))))-------)))).----(((........))).)))).)))))... ( -33.10) >DroEre_CAF1 7381 96 + 1 GUCGGGUGUGGAGCUCGGCUGUACUUUCAGGAUGUAGGAGAACGUCUUGGCAGUGUUAUCUGCA--------AGGA----UAUUCUUGAUUAAGCCCACUCUCACUCA ...(((((.((((...((((.((((.(((((((((......))))))))).)))).......((--------(((.----...)))))....))))..))))))))). ( -31.90) >DroYak_CAF1 7413 96 + 1 AUCGGGAGUGGACUUCGGCAGCACUUUCAGGAUGUAGGAGAAGGUCUUGGCAGUGUUAUCUGCA--------AGGA----UAUUCUCAAUGAAACCCACUCUCACACA ...((((((((..((((...(((((....)).)))..(((((.(((((.((((......)))).--------))))----).)))))..))))..))))))))..... ( -34.20) >DroAna_CAF1 7423 106 + 1 CUCCCCCGUGUCUGGGAGUUUUACUUUUAGGAUGUAGGAGAAUGUCUUUGAACUGCUGUCUGUUAAUUAAAAAGUAAUUGUUUUGUUCUUUAAAUAAAUAUUGU--CU ((((..(((.((((((((.....)))))))))))..))))(((((.((((((..((.......((((((.....))))))....))..))))))...)))))..--.. ( -17.80) >consensus GUCGGGCGUGGACUGUGGCAGCACUUUCAGGAUGUAGGAGAACGUCUUGGAAGUGUUAUCUGCA________AGGA____UUUUCUUGAUUAAACCCACUCUCACACU ...(((.((((..........((((((((((((((......)))))))))))))).................((((.......))))........)))).)))..... (-17.07 = -17.88 + 0.81)

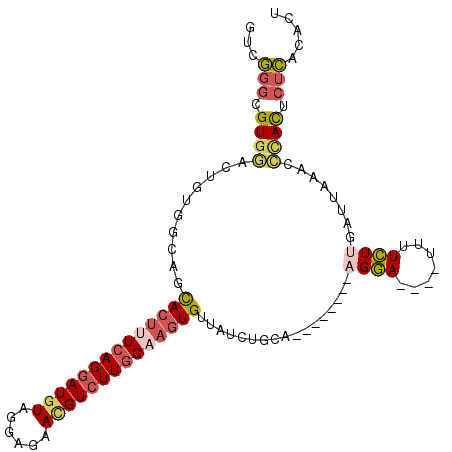

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:09 2006