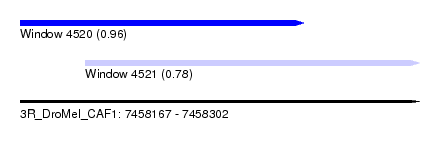
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,458,167 – 7,458,302 |
| Length | 135 |
| Max. P | 0.956633 |

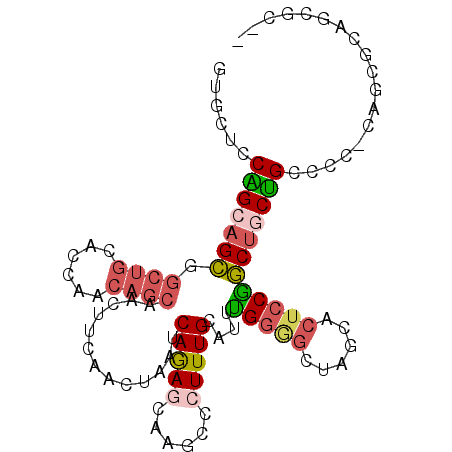
| Location | 7,458,167 – 7,458,263 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 75.53 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -17.52 |
| Energy contribution | -18.17 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
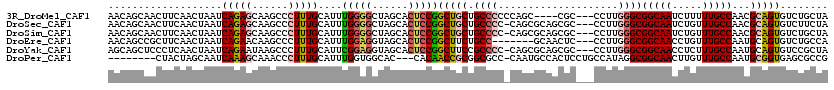
>3R_DroMel_CAF1 7458167 96 + 27905053 GUGCUCCAGCAGCGGCUGCACCAACAGCAACUUCAACUAAUCAGAGCAAGCCCUUUGCAUUUGGGGCUAGCACUCCGGCUGCUGCCCCCCAGC----CGC-- .(((....)))(((((((......(((((.((....((....)).((.((((((........)))))).)).....)).))))).....))))----)))-- ( -33.90) >DroSec_CAF1 12072 99 + 1 GUGCUCCAGCAGCGGCUGCACCAACAGCAACUUCAACUAAUCAGAGCAAGCCCUUUGCAUUUGGGGCUAGCACUCCGGCUGCUGCCCC-CAGCGCAGCGC-- (((((...(((((((((((.......)).................((.((((((........)))))).)).....)))))))))...-.))))).....-- ( -37.90) >DroSim_CAF1 12072 99 + 1 GUGCUCCAGCAGCGGCUGCACCAACAGCAACUUCAACUAAUCAGAGCAAGCCCUUUGCAUUUGGGGCUAGCACUCCGGCUGCUGCCCC-CAGCGCAGCGC-- (((((...(((((((((((.......)).................((.((((((........)))))).)).....)))))))))...-.))))).....-- ( -37.90) >DroEre_CAF1 12920 93 + 1 GUACUCCAGCAGCGGCUGCGCCAACAGCCGCUUCAACUAAUCAGAACAAGCCCUUUGCAUUUGGAGGUAGCACUCCGGCUUCUGCC-------GCAACUC-- ........(.((((((((......)))))))).)....................((((....((((......))))(((....)))-------))))...-- ( -30.90) >DroYak_CAF1 12441 99 + 1 GUGCUCCGGCAGCUGCUGCACCAGCAGCUCCCUCAACUAAUCAGAAUAAGCCCUUUGCAUUCGGAGGUAGCACUCCGGCUUCCGCCCC-CAGCGCAGCGC-- (((((.(((.((((((((...)))))))).))........................((....((((......))))(((....)))..-..))).)))))-- ( -33.80) >DroPer_CAF1 14314 86 + 1 GGGCACCUGCCGUGCCCG------------CUACUAGCAAUCAAAGCAAACCCUUUGCAUUUGGUGGCAC---CACAACCGCGGCGCC-CAAUGCCACUCCU ((((((.....))))))(------------(((((((....(((((......)))))...))))))))..---.........((((..-...))))...... ( -28.20) >consensus GUGCUCCAGCAGCGGCUGCACCAACAGCAACUUCAACUAAUCAGAGCAAGCCCUUUGCAUUUGGGGCUAGCACUCCGGCUGCUGCCCC_CAGCGCAGCGC__ ......(((((((.((((......)))).............(((((......)))))....(((((......)))))))))))).................. (-17.52 = -18.17 + 0.64)


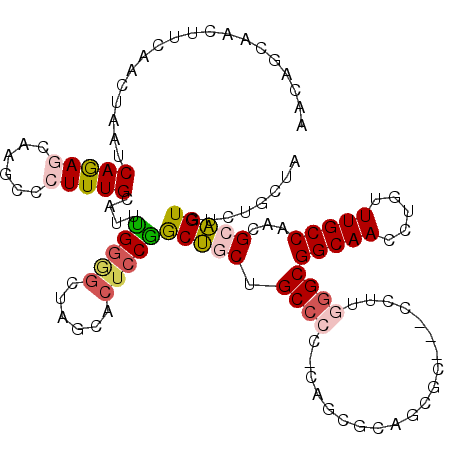
| Location | 7,458,189 – 7,458,302 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.30 |
| Mean single sequence MFE | -38.78 |
| Consensus MFE | -20.56 |
| Energy contribution | -21.23 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7458189 113 + 27905053 AACAGCAACUUCAACUAAUCAGAGCAAGCCCUUUGCAUUUGGGGCUAGCACUCCGGCUGCUGCCCCCCAGC----CGC---CCUUGGGCGGCAAUCUUUUUGCCAACGCAGUGUCUGCUA ...((((................((.((((((........)))))).)).....(((.(((((........----.((---(....)))(((((.....)))))...)))))))))))). ( -40.40) >DroSec_CAF1 12094 116 + 1 AACAGCAACUUCAACUAAUCAGAGCAAGCCCUUUGCAUUUGGGGCUAGCACUCCGGCUGCUGCCCC-CAGCGCAGCGC---CCUUGGGCGGCAAUCUGUUUGCCAACGCAGUGUCUUCUA ....................((((..((((((........)))))).(((((.(((((((.((...-..)))))))((---(....)))(((((.....)))))..)).))))).)))). ( -41.20) >DroSim_CAF1 12094 116 + 1 AACAGCAACUUCAACUAAUCAGAGCAAGCCCUUUGCAUUUGGGGCUAGCACUCCGGCUGCUGCCCC-CAGCGCAGCGC---CCUUGGGCGGCAAUCUGUUUGCCAACGCAGUGUCUGCUA ......................((((((((((........)))))).(((((.(((((((.((...-..)))))))((---(....)))(((((.....)))))..)).))))).)))). ( -44.10) >DroEre_CAF1 12942 110 + 1 AACAGCCGCUUCAACUAAUCAGAACAAGCCCUUUGCAUUUGGAGGUAGCACUCCGGCUUCUGCC-------GCAACUC---CCUUGGGCGGCAACCUGUUUGCCAAUGCAGUGUCUGCCA ..(((.((((.((..............((((.((((....((((......))))(((....)))-------))))...---....))))(((((.....)))))..)).)))).)))... ( -36.00) >DroYak_CAF1 12463 116 + 1 AGCAGCUCCCUCAACUAAUCAGAAUAAGCCCUUUGCAUUCGGAGGUAGCACUCCGGCUUCCGCCCC-CAGCGCAGCGC---CCUUGGGCGGCAACCUCUUUGCCAAUGCAGUGUCCGCUA .(((.......................((((...((...(((((......)))))(((..(((...-..))).)))))---....))))(((((.....)))))..)))(((....))). ( -35.40) >DroPer_CAF1 14332 108 + 1 --------CUACUAGCAAUCAAAGCAAACCCUUUGCAUUUGGUGGCAC---CACAACCGCGGCGCC-CAAUGCCACUCCUGCCAUAGGCGGCAACUUGUUUGCCAAUGCGGUGAGCGCCG --------......((.((((((((((.....)))).)))))).))..---........((((((.-((.(((.......(((...)))(((((.....)))))...))).)).)))))) ( -35.60) >consensus AACAGCAACUUCAACUAAUCAGAGCAAGCCCUUUGCAUUUGGGGCUAGCACUCCGGCUGCUGCCCC_CAGCGCAGCGC___CCUUGGGCGGCAACCUGUUUGCCAACGCAGUGUCUGCUA ...................(((((......)))))....(((((......)))))(((((.((((....................))))(((((.....)))))...)))))........ (-20.56 = -21.23 + 0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:49 2006