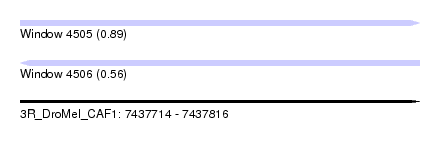
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,437,714 – 7,437,816 |
| Length | 102 |
| Max. P | 0.891950 |

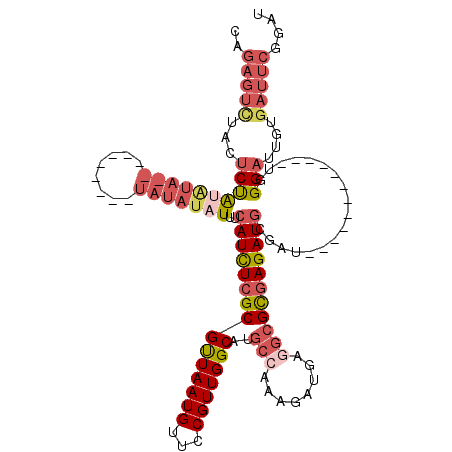
| Location | 7,437,714 – 7,437,816 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.92 |
| Mean single sequence MFE | -23.34 |
| Consensus MFE | -11.18 |
| Energy contribution | -12.46 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7437714 102 + 27905053 AUCCGAAUCCAAAUCCCA------------AUCGCAUCUCGCGCCUCAUCUGUGGCAUGCCAACGGAACAUUAACGCGAGAUGAAAUAUAUAUA------UAUAUAUAGAGUAGACUCUG ..................------------....(((((((((.....(((((((....)).))))).......)))))))))..((((((...------.))))))((((....)))). ( -26.30) >DroSec_CAF1 20501 96 + 1 AUCCGAAUCACAAUCCCA------------AUCGCAUCUCGCGCCUCAUCUUUGGCAUGCCAACGGAACAUUAACGCGAAAUGAAAUAUA------------UAUAUAGAGUAGACUCUG ..................------------....(((.(((((.....(((((((....)))).))).......))))).))).......------------....(((((....))))) ( -17.10) >DroSim_CAF1 19113 96 + 1 AUCCGAAUCACAAUCCCA------------AUCGAAUCUCGCGCCUCAUCUUUGGCAUGCCAACGGAACAUUAACGCGAGAUGAAAUAUA------------UACAUAGAGUAGACUCUG ..................------------.....((((((((.....(((((((....)))).))).......))))))))........------------....(((((....))))) ( -20.40) >DroYak_CAF1 21150 110 + 1 AUCGGAAUCACAAUCCCAAUCCCAAUCCUCAUCUCAUCUCGCGCCC----------GAGCCAACGGAACAUUAGCGCGAGAUGAAAAAUAUAUAUGUAUGUAUAUAUGGAGUAAACUCGG ...(((.......))).....((..........(((((((((((((----------(......))).......)))))))))))...((((((((....)))))))).(((....))))) ( -29.81) >DroAna_CAF1 18658 97 + 1 GUUGGAGAUAGAAGCCCAAUCUUUAUCC-----ACAUCUAACGCUUCAUCUUUGGCAUGCCAACGGCACAUUAACGCGAGAUGAUAUAUAUAA-----------GCCAGAGUA------- ((((((((((((.........)))))).-----...)))))).......(((((((.((((...)))).((((.(....).))))........-----------)))))))..------- ( -23.10) >consensus AUCCGAAUCACAAUCCCA____________AUCGCAUCUCGCGCCUCAUCUUUGGCAUGCCAACGGAACAUUAACGCGAGAUGAAAUAUAUA__________UAUAUAGAGUAGACUCUG ..................................(((((((((((........)))...((...)).........))))))))..................................... (-11.18 = -12.46 + 1.28)

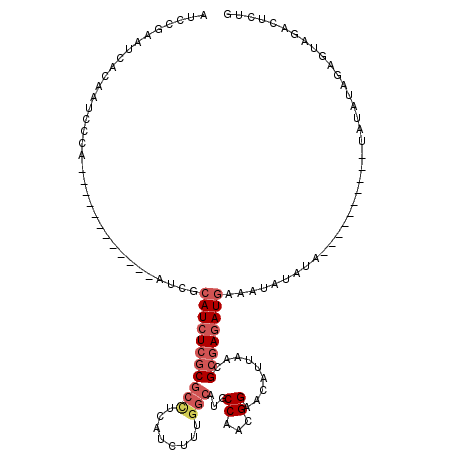
| Location | 7,437,714 – 7,437,816 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.92 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -16.84 |
| Energy contribution | -19.80 |
| Covariance contribution | 2.96 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7437714 102 - 27905053 CAGAGUCUACUCUAUAUAUA------UAUAUAUAUUUCAUCUCGCGUUAAUGUUCCGUUGGCAUGCCACAGAUGAGGCGCGAGAUGCGAU------------UGGGAUUUGGAUUCGGAU ..((((((((((((((((..------..))))))((.(((((((((((((((...)))))))..(((........))))))))))).)).------------.)))...))))))).... ( -31.50) >DroSec_CAF1 20501 96 - 1 CAGAGUCUACUCUAUAUA------------UAUAUUUCAUUUCGCGUUAAUGUUCCGUUGGCAUGCCAAAGAUGAGGCGCGAGAUGCGAU------------UGGGAUUGUGAUUCGGAU ..(((((((.(((.....------------...(((.(((((((((((((((...)))))))..(((........))))))))))).)))------------..))).)).))))).... ( -26.00) >DroSim_CAF1 19113 96 - 1 CAGAGUCUACUCUAUGUA------------UAUAUUUCAUCUCGCGUUAAUGUUCCGUUGGCAUGCCAAAGAUGAGGCGCGAGAUUCGAU------------UGGGAUUGUGAUUCGGAU ..(((((.((...((.(.------------...(((..((((((((((((((...)))))))..(((........))))))))))..)))------------..).)).))))))).... ( -25.20) >DroYak_CAF1 21150 110 - 1 CCGAGUUUACUCCAUAUAUACAUACAUAUAUAUUUUUCAUCUCGCGCUAAUGUUCCGUUGGCUC----------GGGCGCGAGAUGAGAUGAGGAUUGGGAUUGGGAUUGUGAUUCCGAU ((.((((..(((.(((((((......))))))).((((((((((((((...(((.....)))..----------.)))))))))))))).))))))).))((((((((....)))))))) ( -38.60) >DroAna_CAF1 18658 97 - 1 -------UACUCUGGC-----------UUAUAUAUAUCAUCUCGCGUUAAUGUGCCGUUGGCAUGCCAAAGAUGAAGCGUUAGAUGU-----GGAUAAAGAUUGGGCUUCUAUCUCCAAC -------......(((-----------(((........(((.(((((((((((..((((((....)))...)))..))))).)))))-----))))......))))))............ ( -18.74) >consensus CAGAGUCUACUCUAUAUA__________UAUAUAUUUCAUCUCGCGUUAAUGUUCCGUUGGCAUGCCAAAGAUGAGGCGCGAGAUGCGAU____________UGGGAUUGUGAUUCGGAU ..(((((...((((((((((........)))))))..(((((((((((((((...)))))))..(((........)))))))))))..................)))....))))).... (-16.84 = -19.80 + 2.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:34 2006