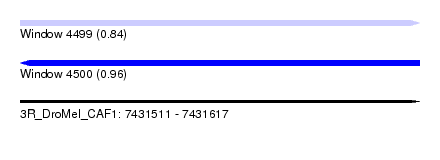
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,431,511 – 7,431,617 |
| Length | 106 |
| Max. P | 0.959641 |

| Location | 7,431,511 – 7,431,617 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 86.82 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -23.86 |
| Energy contribution | -25.30 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7431511 106 + 27905053 GCAGCAGCAGUAGCAGUAGUUGCAAACUCUAAUUUCAAGUGUCUUGCCACUAAUGAGGCCGCCCCUUUUGCCCCUUUUCCUGUAUUUGCCACACUUGCUGCUGCUU--- (((((((((((.((((((((.((((((.((.......)).)).)))).))))..((((..((.......)).)))).........)))).))...)))))))))..--- ( -31.10) >DroSec_CAF1 14364 106 + 1 GCAGCAGCAGUAGCAGUAGUUGCAAACUCUAAUUUCAAGUGUCUUGCCACUAAUGAGGCCGCCCCUUUUGCCGCUUUUCCUGUAUUUGCCACACUUGCUGCAGCUU--- ((((((((((..((((((((.((((((.((.......)).)).)))).))))..(((((.((.......)).)))))..))))..))))......)))))).....--- ( -29.10) >DroEre_CAF1 14289 103 + 1 GAAGUAGCAGCAGCA------GCAAACUCUAAUUUCAAGUGUCUUGCCACUAAUGAGGCCGCCCCUUUUGCCGAUUUUCCUCUAUUUGCCGCACUUUCUGCUGCUGCAC ...(((((((((((.------(((((........(((((((......))))..)))(((..........)))............))))).))......))))))))).. ( -27.80) >DroYak_CAF1 14834 103 + 1 GCAGCAGCAGCAGCA------GCAAACUCUAAUUUCAAGUGUCUUGCUACUAAUGAGGCCGCCCCUUUUGCCACUUUUCCUGAAUCUGCCGCACUUGCUGCUGAUGUGC .((((((((((.(((------((((((.((.......)).)).)))))........(((..........)))...............)).))...))))))))...... ( -26.60) >consensus GCAGCAGCAGCAGCA______GCAAACUCUAAUUUCAAGUGUCUUGCCACUAAUGAGGCCGCCCCUUUUGCCGCUUUUCCUGUAUUUGCCACACUUGCUGCUGCUG___ ...((((((((((((......(((((........(((((((......))))..)))(((..........)))............)))))......)))))))))))).. (-23.86 = -25.30 + 1.44)


| Location | 7,431,511 – 7,431,617 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 86.82 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -26.00 |
| Energy contribution | -26.75 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7431511 106 - 27905053 ---AAGCAGCAGCAAGUGUGGCAAAUACAGGAAAAGGGGCAAAAGGGGCGGCCUCAUUAGUGGCAAGACACUUGAAAUUAGAGUUUGCAACUACUGCUACUGCUGCUGC ---..(((((((((...((((((............(((((..........)))))..((((.((((...((((.......)))))))).)))).))))))))))))))) ( -38.30) >DroSec_CAF1 14364 106 - 1 ---AAGCUGCAGCAAGUGUGGCAAAUACAGGAAAAGCGGCAAAAGGGGCGGCCUCAUUAGUGGCAAGACACUUGAAAUUAGAGUUUGCAACUACUGCUACUGCUGCUGC ---..((.(((((..((((.....))))......((..(((..((..(((..(((..((((..((((...))))..)))))))..)))..))..)))..))))))).)) ( -32.90) >DroEre_CAF1 14289 103 - 1 GUGCAGCAGCAGAAAGUGCGGCAAAUAGAGGAAAAUCGGCAAAAGGGGCGGCCUCAUUAGUGGCAAGACACUUGAAAUUAGAGUUUGC------UGCUGCUGCUACUUC ((((((((((((....(((.((.(((.((((....(((.(......).)))))))))).)).)))((((.((.......)).)))).)------))))))))).))... ( -36.80) >DroYak_CAF1 14834 103 - 1 GCACAUCAGCAGCAAGUGCGGCAGAUUCAGGAAAAGUGGCAAAAGGGGCGGCCUCAUUAGUAGCAAGACACUUGAAAUUAGAGUUUGC------UGCUGCUGCUGCUGC ......((((((((.(((((((((((((.......(.(((..........))).)..((((..((((...))))..))))))))))))------))).)))))))))). ( -38.50) >consensus ___AAGCAGCAGCAAGUGCGGCAAAUACAGGAAAAGCGGCAAAAGGGGCGGCCUCAUUAGUGGCAAGACACUUGAAAUUAGAGUUUGC______UGCUACUGCUGCUGC .....(((((((((...((((((...............(((((.((((...)))).(((((..((((...))))..)))))..)))))......))))))))))))))) (-26.00 = -26.75 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:29 2006