
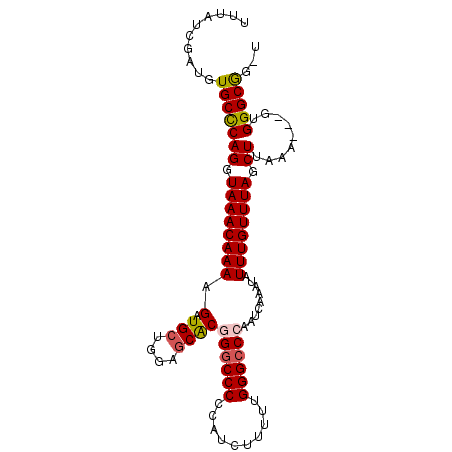
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,355,596 – 7,355,692 |
| Length | 96 |
| Max. P | 0.989643 |

| Location | 7,355,596 – 7,355,692 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -28.41 |
| Consensus MFE | -22.66 |
| Energy contribution | -23.22 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7355596 96 + 27905053 ACCCGCCCAC---UUUAAGGUAAACAAAUAUUUGAUUGGCCCCAAAAAGAUGGGGGCCCGUGCUCCAGCAUCUUUUGUUUACCUGAGCACAACGAUAAA ...((.....---....(((((((((((.........((((((.........)))))).((((....))))..)))))))))))........))..... ( -28.43) >DroSec_CAF1 31616 96 + 1 AACCGCCCAC---UUUAAGCUAAACAAAUAUUUGAUUGGGCCCAAAAAGAUGGGGGCCCGUGCUCCAGCAUCUUUUGUUUACCUGGGCACAUCGAUAAA ....(((((.---.....(.((((((((.........((((((..........))))))((((....))))..)))))))).))))))........... ( -29.10) >DroSim_CAF1 37081 96 + 1 AACCGCCCAC---UUUAAGCUAAACAAAUAUUUGAUUGGGCCCAAAAAGAUGGGGGCCCGUGCUCCAGCAUCUUUUGUUUACCUGGGCACAUCGAUAAA ....(((((.---.....(.((((((((.........((((((..........))))))((((....))))..)))))))).))))))........... ( -29.10) >DroEre_CAF1 35388 92 + 1 ---UGCCCCC---UUCGAGCUAAACAAAUAUUUGAUUGGGCCCAA-AAGAUGGGGGCCUGUGCUCCACCAUCUUUUGUUUACCUGGGCACAUCGAUAAA ---(((((((---.(((((.((......)))))))..))...(((-(((((((((((....)).)).)))))))))).......))))).......... ( -28.30) >DroYak_CAF1 36421 95 + 1 ---CGCCCCCCUUUUCAAGCUAAACAAAUAUUUGAUUGGGCCCAA-AAGAUGGGGGCCUGCGCUUCAGCAUCUUUUGUUUACCUGGGCACAUCGAUAAA ---.(((((((...(((((.((......)))))))..)))..(((-(((((((((((....)))))..))))))))).......))))........... ( -27.10) >consensus A_CCGCCCAC___UUUAAGCUAAACAAAUAUUUGAUUGGGCCCAAAAAGAUGGGGGCCCGUGCUCCAGCAUCUUUUGUUUACCUGGGCACAUCGAUAAA ....((((.........((.((((((((.........((((((..........))))))((((....))))..)))))))).))))))........... (-22.66 = -23.22 + 0.56)

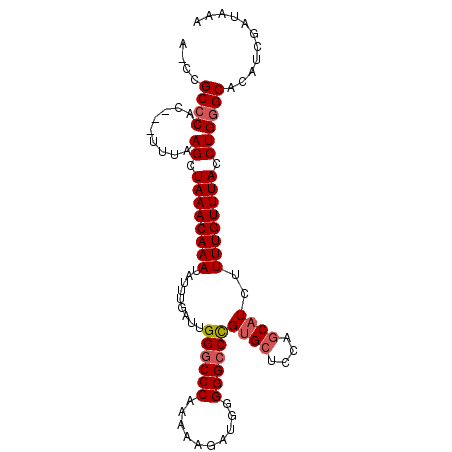

| Location | 7,355,596 – 7,355,692 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -23.42 |
| Energy contribution | -23.74 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7355596 96 - 27905053 UUUAUCGUUGUGCUCAGGUAAACAAAAGAUGCUGGAGCACGGGCCCCCAUCUUUUUGGGGCCAAUCAAAUAUUUGUUUACCUUAAA---GUGGGCGGGU ....(((((.((((.(((((((((((...(((....)))..((((((.........)))))).........)))))))))))...)---)))))))).. ( -31.80) >DroSec_CAF1 31616 96 - 1 UUUAUCGAUGUGCCCAGGUAAACAAAAGAUGCUGGAGCACGGGCCCCCAUCUUUUUGGGCCCAAUCAAAUAUUUGUUUAGCUUAAA---GUGGGCGGUU ..........(((((((.((((((((...(((....))).((((((..........)))))).........)))))))).).....---.))))))... ( -30.10) >DroSim_CAF1 37081 96 - 1 UUUAUCGAUGUGCCCAGGUAAACAAAAGAUGCUGGAGCACGGGCCCCCAUCUUUUUGGGCCCAAUCAAAUAUUUGUUUAGCUUAAA---GUGGGCGGUU ..........(((((((.((((((((...(((....))).((((((..........)))))).........)))))))).).....---.))))))... ( -30.10) >DroEre_CAF1 35388 92 - 1 UUUAUCGAUGUGCCCAGGUAAACAAAAGAUGGUGGAGCACAGGCCCCCAUCUU-UUGGGCCCAAUCAAAUAUUUGUUUAGCUCGAA---GGGGGCA--- ........(((.(((.(((...((((((((((.((.((....)))))))))))-))).)))..........((((.......))))---))).)))--- ( -32.00) >DroYak_CAF1 36421 95 - 1 UUUAUCGAUGUGCCCAGGUAAACAAAAGAUGCUGAAGCGCAGGCCCCCAUCUU-UUGGGCCCAAUCAAAUAUUUGUUUAGCUUGAAAAGGGGGGCG--- ........(((((...((((.........))))...))))).((((((...((-(..(((.....(((....)))....)))..)))..)))))).--- ( -30.00) >consensus UUUAUCGAUGUGCCCAGGUAAACAAAAGAUGCUGGAGCACGGGCCCCCAUCUUUUUGGGCCCAAUCAAAUAUUUGUUUAGCUUAAA___GUGGGCGG_U ..........(((((((.((((((((.(.(((....))))((((((..........)))))).........)))))))).)).........)))))... (-23.42 = -23.74 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:55 2006