
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
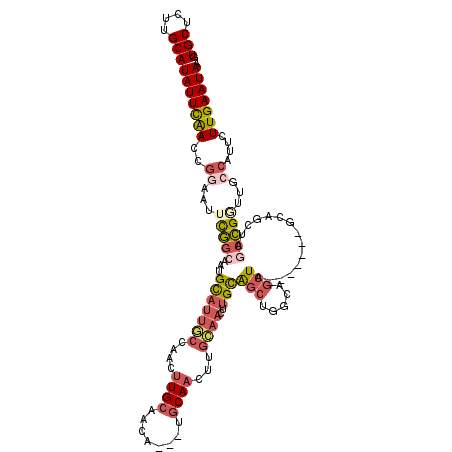
| Location | 7,353,145 – 7,353,294 |
| Length | 149 |
| Max. P | 0.988149 |

| Location | 7,353,145 – 7,353,255 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.15 |
| Mean single sequence MFE | -33.42 |
| Consensus MFE | -15.99 |
| Energy contribution | -17.33 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
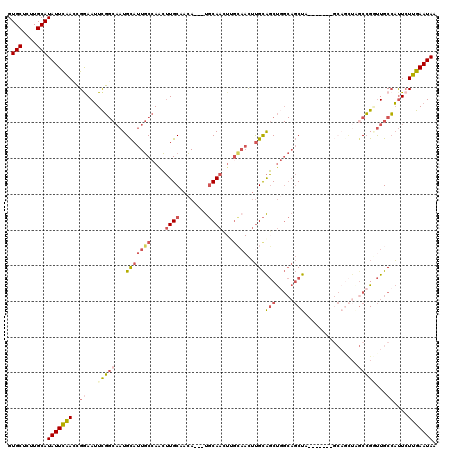
>3R_DroMel_CAF1 7353145 110 + 27905053 GUGCUCGUGCAUAUUCAAACGGAAUUCGGCAAUGCAUUGCCAACUUGCAACA---UGCAACUCGCAACUUGCAGCUGGCAGCUA-------GCAGCUAGCCGGUUGCCAUUCUUGAAUAA .(((....)))((((((...(((((..((((.((((((((....((((....---.))))...))))..))))(((((((((..-------...))).))))))))))))))))))))). ( -36.80) >DroSec_CAF1 29171 103 + 1 GUGCUCUUGCAUAUUUGAUCGGAAUUCGGCAAUGCAUUGCCAACUUGCAACA---UGCAA-------CUUGCAGCUGGCAGCUA-------GCAUCUAGCCGGUUGCCAUUCUUGAAUAA ((((....))))...........(((((((((((((((((......)))..)---)))).-------.))))...(((((((..-------((.....))..)))))))....))))).. ( -30.80) >DroSim_CAF1 34637 110 + 1 GUGCUCUUGCAUAUUCGACCGGAAUUCGGCAAUGCAUUGCCAACUUGCAACA---UGCAACUUGCAACUUGCAGCUGGCAGCUA-------GCAGCUAGCCGGUUGCCAUUCUUGAAUAA .(((....)))((((((...(((((..(((((....)))))...((((((..---......))))))...((((((((((((..-------...))).))))))))).))))))))))). ( -37.10) >DroEre_CAF1 33004 103 + 1 GUGCUCUUGCAUAUUCAACCGGAAUUCGGCAAUGCAUUGCCAACUUGCCACA---UGCAACUUGCAACUUGCAGCUGGCAGCUUG--------------CCGUUUGCCAUUCUUGAAUAA .(((....)))(((((((..((....((((((.((..(((((.((.((....---(((.....)))....)))).))))))))))--------------)))....))....))))))). ( -33.40) >DroYak_CAF1 33956 117 + 1 GUGCUCCUGCAUAUUCAACCGAAAUUCGGCAAUGCAUUGCCAACUUGCCACA---UGCAACUUGCAACUUGCAGCUUGCAGCUUGCAACUUGCAGCUGGCCGUUUGCCAUUCUUGAAUAA .(((....)))(((((((..(((....(((((....))))).....((((..---(((((.((((((.((((.....)))).)))))).)))))..)))).........)))))))))). ( -38.10) >DroAna_CAF1 31570 101 + 1 GUGCACUUGCAUAUUCAACGGAAAAUUU---UUGUAUUUC-AGGAUGCUAGAAUCCUCAAAUCGGAAUGCGUGUCC-----CU----------CGUUUGCAGACUGCAUCUAUUGAAUAA .(((....)))(((((((..((....((---(((.((((.-(((((......))))).)))))))))((((.(((.-----..----------........)))))))))..))))))). ( -24.30) >consensus GUGCUCUUGCAUAUUCAACCGGAAUUCGGCAAUGCAUUGCCAACUUGCAACA___UGCAACUUGCAACUUGCAGCUGGCAGCUA_______GCAGCUAGCCGGUUGCCAUUCUUGAAUAA .(((....)))(((((((..((...(((((...(((((((....((((........))))...))))..)))(((.....)))...............)))))...))....))))))). (-15.99 = -17.33 + 1.34)

| Location | 7,353,145 – 7,353,255 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.15 |
| Mean single sequence MFE | -35.65 |
| Consensus MFE | -15.86 |
| Energy contribution | -18.38 |
| Covariance contribution | 2.52 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7353145 110 - 27905053 UUAUUCAAGAAUGGCAACCGGCUAGCUGC-------UAGCUGCCAGCUGCAAGUUGCGAGUUGCA---UGUUGCAAGUUGGCAAUGCAUUGCCGAAUUCCGUUUGAAUAUGCACGAGCAC .(((((((..((((....((((....(((-------....((((((((((((..(((.....)))---..)))).))))))))..)))..))))....)))))))))))(((....))). ( -39.10) >DroSec_CAF1 29171 103 - 1 UUAUUCAAGAAUGGCAACCGGCUAGAUGC-------UAGCUGCCAGCUGCAAG-------UUGCA---UGUUGCAAGUUGGCAAUGCAUUGCCGAAUUCCGAUCAAAUAUGCAAGAGCAC ........((.(((....((((..(((((-------....((((((((((((.-------.....---..)))).))))))))..)))))))))....))).)).....(((....))). ( -32.80) >DroSim_CAF1 34637 110 - 1 UUAUUCAAGAAUGGCAACCGGCUAGCUGC-------UAGCUGCCAGCUGCAAGUUGCAAGUUGCA---UGUUGCAAGUUGGCAAUGCAUUGCCGAAUUCCGGUCGAAUAUGCAAGAGCAC .(((((.......(((...((((((...)-------)))))(((((((((((..(((.....)))---..)))).)))))))..)))...((((.....)))).)))))(((....))). ( -38.10) >DroEre_CAF1 33004 103 - 1 UUAUUCAAGAAUGGCAAACGG--------------CAAGCUGCCAGCUGCAAGUUGCAAGUUGCA---UGUGGCAAGUUGGCAAUGCAUUGCCGAAUUCCGGUUGAAUAUGCAAGAGCAC .(((((((...(((....(((--------------(((((((((((((((....(((.....)))---....)).))))))))..)).))))))....))).)))))))(((....))). ( -38.90) >DroYak_CAF1 33956 117 - 1 UUAUUCAAGAAUGGCAAACGGCCAGCUGCAAGUUGCAAGCUGCAAGCUGCAAGUUGCAAGUUGCA---UGUGGCAAGUUGGCAAUGCAUUGCCGAAUUUCGGUUGAAUAUGCAGGAGCAC ...(((.....((.(((...((((..(((((.((((((..(((.....)))..)))))).)))))---..))))...))).)).(((((.((((.....)))).....)))))))).... ( -42.50) >DroAna_CAF1 31570 101 - 1 UUAUUCAAUAGAUGCAGUCUGCAAACG----------AG-----GGACACGCAUUCCGAUUUGAGGAUUCUAGCAUCCU-GAAAUACAA---AAAUUUUCCGUUGAAUAUGCAAGUGCAC .((((((((.((((((((((.((((..----------.(-----(((......))))..)))).)))))...)))))..-((((.....---....)))).))))))))(((....))). ( -22.50) >consensus UUAUUCAAGAAUGGCAACCGGCUAGCUGC_______UAGCUGCCAGCUGCAAGUUGCAAGUUGCA___UGUUGCAAGUUGGCAAUGCAUUGCCGAAUUCCGGUUGAAUAUGCAAGAGCAC .((((((((((((((((.(((....)))..........(((((((((((((...(((.....)))...)))....))))))))..)).)))))..))))...)))))))(((....))). (-15.86 = -18.38 + 2.52)

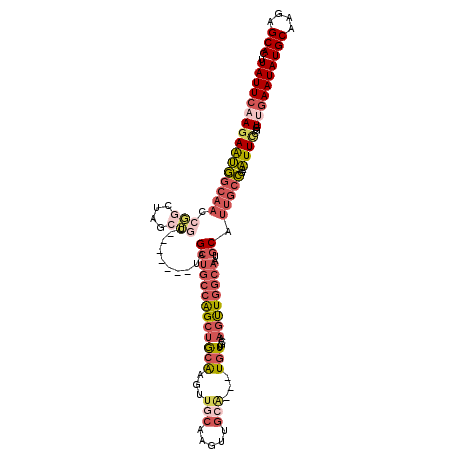

| Location | 7,353,185 – 7,353,294 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.95 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -14.52 |
| Energy contribution | -16.17 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.47 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7353185 109 + 27905053 CAACUUGCAACA---UGCAACUCGCAACUUGCAGCUGGCAGCUA-------GCAGCUAGCCGGUUGCCAUUCUUGAAUAAUUAACCGAAUCCUGAUGGUCCUCGAAUGGGCAAUCAGCU ....((((((..---(((.....)))..)))))).(((((((..-------((.....))..)))))))......................(((((.((((......)))).))))).. ( -33.70) >DroSec_CAF1 29211 102 + 1 CAACUUGCAACA---UGCAA-------CUUGCAGCUGGCAGCUA-------GCAUCUAGCCGGUUGCCAUUCUUGAAUAAUUAACCGCAUCCUGAUGGUCCUCGAAUGGGCAAUCAGCU .....(((....---(((..-------...)))..(((((((..-------((.....))..))))))).................)))..(((((.((((......)))).))))).. ( -31.70) >DroSim_CAF1 34677 109 + 1 CAACUUGCAACA---UGCAACUUGCAACUUGCAGCUGGCAGCUA-------GCAGCUAGCCGGUUGCCAUUCUUGAAUAAUUAACCGCAUCCUGAUGGUCCUCGAAUGGGCAAUCAGCU .....(((....---(((.....)))....((((((((((((..-------...))).)))))))))...................)))..(((((.((((......)))).))))).. ( -36.20) >DroEre_CAF1 33044 102 + 1 CAACUUGCCACA---UGCAACUUGCAACUUGCAGCUGGCAGCUUG--------------CCGUUUGCCAUUCUUGAAUAAUUAACCGAAUCCUGAUGGUCCUCGAAUGGGCAAUCAGCU (((.((((((..---(((((........)))))..)))))).)))--------------................................(((((.((((......)))).))))).. ( -25.30) >DroYak_CAF1 33996 116 + 1 CAACUUGCCACA---UGCAACUUGCAACUUGCAGCUUGCAGCUUGCAACUUGCAGCUGGCCGUUUGCCAUUCUUGAAUAAUUAACCGAACCCUGAUGGUCCUCGAAUGGGCAAUCAGCU ......((((..---(((((.((((((.((((.....)))).)))))).)))))..)))).(((((...................))))).(((((.((((......)))).))))).. ( -35.81) >DroAna_CAF1 31607 103 + 1 -AGGAUGCUAGAAUCCUCAAAUCGGAAUGCGUGUCC-----CU----------CGUUUGCAGACUGCAUCUAUUGAAUAAUUAACCGAUUUCUGAUUGUUUUCGAAUGGGCAAUCAACU -(((((......))))).(((((((..((((.(((.-----..----------........)))))))((....))........))))))).(((((((((......)))))))))... ( -24.30) >consensus CAACUUGCAACA___UGCAACUUGCAACUUGCAGCUGGCAGCUA_______GCAGCUAGCCGGUUGCCAUUCUUGAAUAAUUAACCGAAUCCUGAUGGUCCUCGAAUGGGCAAUCAGCU .....(((........)))...........((((.(((((((............))).)))).))))........................(((((.((((......)))).))))).. (-14.52 = -16.17 + 1.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:53 2006