
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,337,315 – 7,337,452 |
| Length | 137 |
| Max. P | 0.999934 |

| Location | 7,337,315 – 7,337,412 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 93.14 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -20.78 |
| Energy contribution | -20.50 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
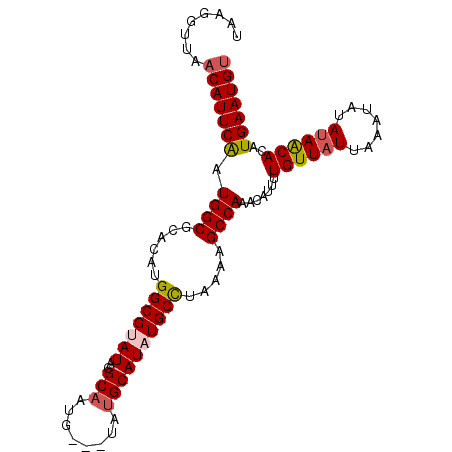
>3R_DroMel_CAF1 7337315 97 + 27905053 GUUAUCUAGGAAUAGAAAGCUUUAUGCGUUAUAAGCUUCAACAUUCAUGUGUUAUAUAUUUAAUAACAAAAUGUUUGGCUUUUAGGCAUAUGCAUA---C ....((((....))))..((..(((((.(((.(((((..((((((....((((((.......)))))).)))))).))))).)))))))).))...---. ( -22.90) >DroSec_CAF1 13521 100 + 1 UUUAUCUAGGACUAGAAAGCUUUAUGCGUUGUAAGCUUCAACAUUCAUGUGUUGUAUAUUUAAUAACAAAAUGGUUGGCUUUUAGGCAUAUGCAUACUAC ....((((....))))..((..(((((.(...(((((....((((....((((((.......)))))).))))...)))))..).))))).))....... ( -21.50) >DroSim_CAF1 18093 100 + 1 UUUAUCUAGGACUAGAAAGCUUUAUGCGUUGUAAGCUUCAACAUUCAUGUGCUAUAUAUUUAAUAACAAAAUGGUUGGCUUUUAGGCAUAUGCAUACUAC ....((((....))))..((..(((((.(...(((((....((((..(((...............))).))))...)))))..).))))).))....... ( -18.06) >DroEre_CAF1 17336 97 + 1 UUUAUCUGGGACUAGAAAGCUUUAUGCGUUGUAAGCUUCAACAUUCAUGUGUUAUAUAUUUAAUAACAAAAUGUUUGGCUUUUAAGCAUAUGCAUA---U ....((((....))))..((..(((((.(((.(((((..((((((....((((((.......)))))).)))))).))))).)))))))).))...---. ( -22.40) >DroYak_CAF1 17935 97 + 1 UUUAUCUGGGACUAGAAAGCUUUAUGCGUUGUAAGCUUCAGCAUUCAUGUGUUAUAUAUUUAAUAACAAAAUGUUUGGCUUUUAAGCAUAUGCCUA---C ....((((....))))..((..(((((.(((.(((((..((((((....((((((.......)))))).)))))).))))).)))))))).))...---. ( -21.50) >consensus UUUAUCUAGGACUAGAAAGCUUUAUGCGUUGUAAGCUUCAACAUUCAUGUGUUAUAUAUUUAAUAACAAAAUGUUUGGCUUUUAGGCAUAUGCAUA___C ....((((....))))..((..(((((.(((.(((((..((((((....((((((.......)))))).)))))).))))).)))))))).))....... (-20.78 = -20.50 + -0.28)



| Location | 7,337,355 – 7,337,452 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 90.92 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -20.72 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.998286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7337355 97 + 27905053 ACAUUCAUGUGUUAUAUAUUUAAUAACAAAAUGUUUGGCUUUUAGGCAUAUGCAUA---CAUUGCGUAUACGCCAUGUACGCCAUCGAAUGUUAAACUUA ((((((...((((((.......)))))).......((((...((((((((((((..---...)))))))..))).))...))))..))))))........ ( -24.40) >DroSec_CAF1 13561 100 + 1 ACAUUCAUGUGUUGUAUAUUUAAUAACAAAAUGGUUGGCUUUUAGGCAUAUGCAUACUACAUUGCGUAUUCGCCAUGUGCGCCAUUGAAUGUUAACCUUA ((((((...((((((.......)))))).((((((..((.....((((((((((........)))))))..)))....))))))))))))))........ ( -26.00) >DroSim_CAF1 18133 100 + 1 ACAUUCAUGUGCUAUAUAUUUAAUAACAAAAUGGUUGGCUUUUAGGCAUAUGCAUACUACAUUGCGUAUUCGCCAUGUGCGCCAUUGAAUGUUAACCUUA (((((((.((((.((((......((((......)))).......((((((((((........)))))))..)))))))))))...)))))))........ ( -24.90) >DroEre_CAF1 17376 97 + 1 ACAUUCAUGUGUUAUAUAUUUAAUAACAAAAUGUUUGGCUUUUAAGCAUAUGCAUA---UAUUGCGUAUACGCCAUGUGCGCCAUUGAAUGUGAACCCUA (((((((..((((((.......)))))).......((((......(.(((((((..---...))))))).)((.....)))))).)))))))........ ( -23.00) >DroYak_CAF1 17975 97 + 1 GCAUUCAUGUGUUAUAUAUUUAAUAACAAAAUGUUUGGCUUUUAAGCAUAUGCCUA---CAUUGCGUAUACGCCAUGUGUGCCAUCGAAUGUUGGCCUUA ((((((...((((((.......)))))).......((((......((....)).((---(((.(((....))).))))).))))..))))))........ ( -22.60) >consensus ACAUUCAUGUGUUAUAUAUUUAAUAACAAAAUGUUUGGCUUUUAGGCAUAUGCAUA___CAUUGCGUAUACGCCAUGUGCGCCAUUGAAUGUUAACCUUA ((((((...((((((.......)))))).......((((.....((((((((((........)))))))..)))......))))..))))))........ (-20.72 = -21.20 + 0.48)



| Location | 7,337,355 – 7,337,452 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 90.92 |
| Mean single sequence MFE | -23.44 |
| Consensus MFE | -22.04 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.65 |
| SVM RNA-class probability | 0.999934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7337355 97 - 27905053 UAAGUUUAACAUUCGAUGGCGUACAUGGCGUAUACGCAAUG---UAUGCAUAUGCCUAAAAGCCAAACAUUUUGUUAUUAAAUAUAUAACACAUGAAUGU ........(((((((.((((......(((((((..(((...---..)))))))))).....)))).......((((((.......))))))..))))))) ( -25.50) >DroSec_CAF1 13561 100 - 1 UAAGGUUAACAUUCAAUGGCGCACAUGGCGAAUACGCAAUGUAGUAUGCAUAUGCCUAAAAGCCAACCAUUUUGUUAUUAAAUAUACAACACAUGAAUGU ........(((((((.((((((((((.(((....))).)))).))..((....))......))))......((((..........))))....))))))) ( -20.30) >DroSim_CAF1 18133 100 - 1 UAAGGUUAACAUUCAAUGGCGCACAUGGCGAAUACGCAAUGUAGUAUGCAUAUGCCUAAAAGCCAACCAUUUUGUUAUUAAAUAUAUAGCACAUGAAUGU ........(((((((.((((((((((.(((....))).)))).))..((....))......)))).......((((((.......))))))..))))))) ( -23.30) >DroEre_CAF1 17376 97 - 1 UAGGGUUCACAUUCAAUGGCGCACAUGGCGUAUACGCAAUA---UAUGCAUAUGCUUAAAAGCCAAACAUUUUGUUAUUAAAUAUAUAACACAUGAAUGU ........(((((((.((((......(((((((..(((...---..)))))))))).....)))).......((((((.......))))))..))))))) ( -24.30) >DroYak_CAF1 17975 97 - 1 UAAGGCCAACAUUCGAUGGCACACAUGGCGUAUACGCAAUG---UAGGCAUAUGCUUAAAAGCCAAACAUUUUGUUAUUAAAUAUAUAACACAUGAAUGC .........((((((.((((..((((.(((....))).)))---)((((....))))....)))).......((((((.......))))))..)))))). ( -23.80) >consensus UAAGGUUAACAUUCAAUGGCGCACAUGGCGUAUACGCAAUG___UAUGCAUAUGCCUAAAAGCCAAACAUUUUGUUAUUAAAUAUAUAACACAUGAAUGU ........(((((((.((((......(((((((..(((........)))))))))).....)))).......((((((.......))))))..))))))) (-22.04 = -22.40 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:44 2006