
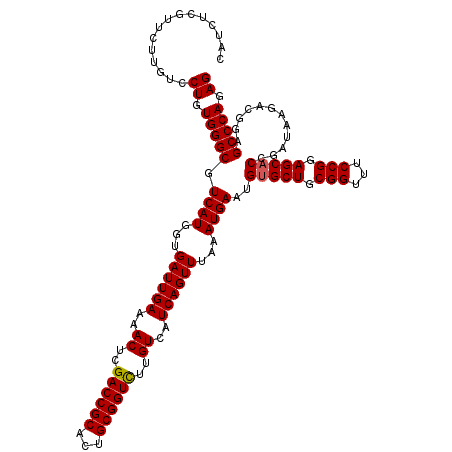
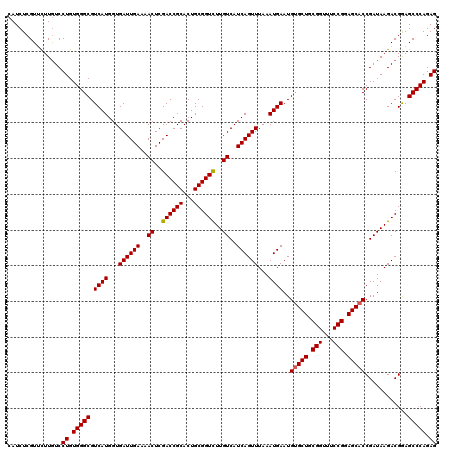
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,326,420 – 7,326,578 |
| Length | 158 |
| Max. P | 0.998343 |

| Location | 7,326,420 – 7,326,538 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.12 |
| Mean single sequence MFE | -32.66 |
| Consensus MFE | -32.46 |
| Energy contribution | -32.38 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7326420 118 + 27905053 --CCCCCCAUCCCCCUGCCAAACACUCGCAAGGACAUCUUCAUCUCGUUCUUGUCCUGUGGGCGUCAUGGUGAUUGAAAACUCGACCGCACUGCGGUCUUGUCAUCAGUUUAAAUGAAUG --..............((((.((.((((((.(((((....(.....)....))))))))))).))..))))((((((..((..((((((...))))))..))..)))))).......... ( -34.50) >DroSec_CAF1 2753 120 + 1 ACCUCCCCUCUCCCCUGCCAAACACUCGGAAGGACAUCUUCAUCUCGUUCUUGUCCUGUGGGCGUCAUGGUGAUUGAAAACUCGACCGCACUGCGGUUUUGUCAUCAGUUUAAAUGAAUG .....(((.((((..((.....))...)))((((((....(.....)....))))))).)))..((((...((((((..((..((((((...))))))..))..))))))...))))... ( -29.70) >DroSim_CAF1 6511 120 + 1 ACCCCCCCUCUCCCCUGCCAAACACUCGCAAGGACAUCUUCAUCUCGUUUUUAUCCUGUGGGCGUCAUGGUGAUUGAAAACUCGACCGCACUGCGGUCUUGUCAUCAGUUUAAAUGAAUG ................((((.((.((((((.(((......(.....)......))))))))).))..))))((((((..((..((((((...))))))..))..)))))).......... ( -29.10) >DroEre_CAF1 6462 116 + 1 ---UCCCCA-CCCCCUGCCAAACACUCGCAAGGACAUCUUCAUCUCGUGCUUGUCCUGUGGGCGUCAUGGUGAUUGAAAACUCGACCGCACUGCGGUCUUGUCAUCAGUUUAAAUGAAUG ---..((((-(....(((.........)))((((((....(((...)))..)))))))))))..((((...((((((..((..((((((...))))))..))..))))))...))))... ( -34.80) >DroYak_CAF1 6655 116 + 1 ---UCCCCC-CUCACUACCAAACACUCACAAGGACAUCUUCAUCUCGUUCUUGUCCUGUGGGCGUCAUGGUGAUUGAAAACUCGACCGCACUGCGGUCUUGUCAUCAGUUUAAAUGAAUG ---......-.(((..((((.((.((((((.(((((....(.....)....))))))))))).))..))))((((((..((..((((((...))))))..))..))))))....)))... ( -35.20) >consensus __CUCCCCA_CCCCCUGCCAAACACUCGCAAGGACAUCUUCAUCUCGUUCUUGUCCUGUGGGCGUCAUGGUGAUUGAAAACUCGACCGCACUGCGGUCUUGUCAUCAGUUUAAAUGAAUG ................((((.((.((((((.(((((....(.....)....))))))))))).))..))))((((((..((..((((((...))))))..))..)))))).......... (-32.46 = -32.38 + -0.08)



| Location | 7,326,458 – 7,326,578 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -42.24 |
| Energy contribution | -42.28 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7326458 120 + 27905053 CAUCUCGUUCUUGUCCUGUGGGCGUCAUGGUGAUUGAAAACUCGACCGCACUGCGGUCUUGUCAUCAGUUUAAAUGAAUGUGCUGCGGUUUCCGGAGCACCGAUAAGACGGAGCCCAGAG ...............((.(((((.((((...((((((..((..((((((...))))))..))..))))))...))))..(((((.(((...))).)))))............))))).)) ( -43.50) >DroSec_CAF1 2793 120 + 1 CAUCUCGUUCUUGUCCUGUGGGCGUCAUGGUGAUUGAAAACUCGACCGCACUGCGGUUUUGUCAUCAGUUUAAAUGAAUGAGCUGCGGUUUCCGGAGCACCGAUAAGACGAAGCCCAGAG ...............((.(((((.((((...((((((..((..((((((...))))))..))..))))))...))))....(((.(((...))).)))..............))))).)) ( -38.50) >DroSim_CAF1 6551 120 + 1 CAUCUCGUUUUUAUCCUGUGGGCGUCAUGGUGAUUGAAAACUCGACCGCACUGCGGUCUUGUCAUCAGUUUAAAUGAAUGUGCUGCGGUUUCCGGAGCACCGAUAAGACGAAGCCCAGAG ...............((.(((((.((((...((((((..((..((((((...))))))..))..))))))...))))..(((((.(((...))).)))))............))))).)) ( -43.50) >DroEre_CAF1 6498 120 + 1 CAUCUCGUGCUUGUCCUGUGGGCGUCAUGGUGAUUGAAAACUCGACCGCACUGCGGUCUUGUCAUCAGUUUAAAUGAAUGUGCUGCGGUUUCCGGAGCACCGAUAAGACGGAGCCCAGAG ...............((.(((((.((((...((((((..((..((((((...))))))..))..))))))...))))..(((((.(((...))).)))))............))))).)) ( -43.50) >DroYak_CAF1 6691 120 + 1 CAUCUCGUUCUUGUCCUGUGGGCGUCAUGGUGAUUGAAAACUCGACCGCACUGCGGUCUUGUCAUCAGUUUAAAUGAAUGUGCUGCGGUUUCCGGAGCACCGAUAAGACGGAGCCCAGAG ...............((.(((((.((((...((((((..((..((((((...))))))..))..))))))...))))..(((((.(((...))).)))))............))))).)) ( -43.50) >consensus CAUCUCGUUCUUGUCCUGUGGGCGUCAUGGUGAUUGAAAACUCGACCGCACUGCGGUCUUGUCAUCAGUUUAAAUGAAUGUGCUGCGGUUUCCGGAGCACCGAUAAGACGGAGCCCAGAG ...............((.(((((.((((...((((((..((..((((((...))))))..))..))))))...))))..(((((.(((...))).)))))............))))).)) (-42.24 = -42.28 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:35 2006