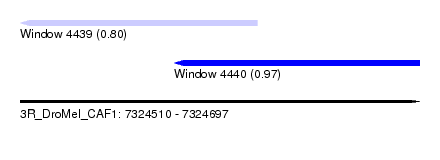
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,324,510 – 7,324,697 |
| Length | 187 |
| Max. P | 0.966848 |

| Location | 7,324,510 – 7,324,621 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -24.60 |
| Energy contribution | -24.44 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.797050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7324510 111 - 27905053 AAUUUAACAAGCGGCGACAUUUUGCCAGCCCCUC-AUCCAUCUCUUUUAGCCCUAAGUGAAUUCUG-------UUUUCAUCGCUGGCUGU-CGGUGAAUAGGGGUGCAGAAUUAUGAAAA ..........(((((((....))))).((((((.-((.((((.....(((((....(((((.....-------..)))))....))))).-.)))).))))))))))............. ( -31.00) >DroSec_CAF1 835 118 - 1 AACUUAACAAGCGGCGACAUUUUGCCAGCCCCUC-AUCCAUCUCUUUUAGCCCUAAGUGAAUUCUGUUUUCUGUUUUCAUCGCUGGCUGU-CGGUGAAUAGGGGUGCAGAAUUAUGAAAA ..........(((((((....))))).((((((.-((.((((.....(((((....(((((..(........)..)))))....))))).-.)))).))))))))))............. ( -32.20) >DroSim_CAF1 4591 118 - 1 AAUUUAACAAGCGGCGACAUUUUGCCAGCCCCUC-AUCCAUCUCUCUUAGCCCUAAGUGAAUUCUGUUUUCUGUUUUCAUCGCUGGCUGU-CGGUGAAUAGGGGUGCAAAAUUAUGAAAA ..........(((((((....))))).((((((.-((.((((.....(((((....(((((..(........)..)))))....))))).-.)))).))))))))))............. ( -32.20) >DroEre_CAF1 4580 112 - 1 AAUUUAACAAGCGGCGACAUUUUGCCAGCCCCUCCAUCAUUCUCCUUUAGCCCAAAGUGAAUUCUG-------UUUUCAUCGCUGGUUUU-CGGUGAAUAGGGGUGCAAAAUUAUGAAAA ..........(((((((....))))).((((((.....((((.((...((((....(((((.....-------..)))))....))))..-.)).))))))))))))............. ( -29.80) >DroYak_CAF1 4705 112 - 1 AAUUUAACAAGCGGCGACAUUUUGCCAGCCCCUC-AUCCAUCUGUUUUAGCCCUAAGUGAAUUCUG-------UUUUCAUCGCUGUUUUGCCAGUGAAUAGGGGUGCAAAAUUAUGAAAA ..........(((((((....))))).((((((.-......................((((.....-------..))))((((((......))))))..))))))))............. ( -26.20) >consensus AAUUUAACAAGCGGCGACAUUUUGCCAGCCCCUC_AUCCAUCUCUUUUAGCCCUAAGUGAAUUCUG_______UUUUCAUCGCUGGCUGU_CGGUGAAUAGGGGUGCAAAAUUAUGAAAA ..........(((((((....))))).((((((........................((((..............))))((((((......))))))..))))))))............. (-24.60 = -24.44 + -0.16)


| Location | 7,324,582 – 7,324,697 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.81 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -24.11 |
| Energy contribution | -24.15 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7324582 115 - 27905053 CGAGCGGGCGC----CUCAGUUGGGAUAGACCACCCACGUCGCUCAUAUUAUUUUACACUAACAAGCCAAACAUUCCUUAAAUUUAACAAGCGGCGACAUUUUGCCAGCCCCUC-AUCCA .(((.((((..----......((((........))))(((((((......((((.........................))))......)))))))...........)))))))-..... ( -26.41) >DroSec_CAF1 914 115 - 1 CGAGCGGGCGC----CUGAGUUGGGAUAGACCACCCACGUCGCUCAUAUUAUUUUACACUAACAAGCCAAACAUUCCUUAAACUUAACAAGCGGCGACAUUUUGCCAGCCCCUC-AUCCA .(((.((((((----.((((((((((..(((.......)))(((....................))).......))))..))))))....))(((((....))))).)))))))-..... ( -28.95) >DroSim_CAF1 4670 115 - 1 CGAGCGGGCGC----CUGAGUUGGGAUAGACCACCCACGUCGCUCAUAUUAUUUUACACUAACAAGCCAAACAUUCCUUAAAUUUAACAAGCGGCGACAUUUUGCCAGCCCCUC-AUCCA .(((.((((..----.(((((((((........))))....)))))..............................................(((((....))))).)))))))-..... ( -28.20) >DroEre_CAF1 4652 120 - 1 CGAGCGGGUGAACGCCUGACUUGGGAUAGACCACCCACGUCGCUCAUAUUAUUUUACACUAACAAGCCAAACAUUCCUUAAAUUUAACAAGCGGCGACAUUUUGCCAGCCCCUCCAUCAU .((((((((....))))(((.((((........)))).))))))).............................................(((((((....))))).))........... ( -27.70) >DroYak_CAF1 4778 115 - 1 CGAGCGGGCGUUCG----UGUUGGGAUAGACCACCCACGUCGCUCAUAUUAUUUUACACUAACAAGCCAAACAUUCCUUAAAUUUAACAAGCGGCGACAUUUUGCCAGCCCCUC-AUCCA .(((.(((((((.(----(((((((...(((.......))).)))).........)))).))).............................(((((....))))).)))))))-..... ( -28.80) >consensus CGAGCGGGCGC____CUGAGUUGGGAUAGACCACCCACGUCGCUCAUAUUAUUUUACACUAACAAGCCAAACAUUCCUUAAAUUUAACAAGCGGCGACAUUUUGCCAGCCCCUC_AUCCA .(((.((((............((((........))))(((((((......((((.........................))))......)))))))...........)))))))...... (-24.11 = -24.15 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:30 2006