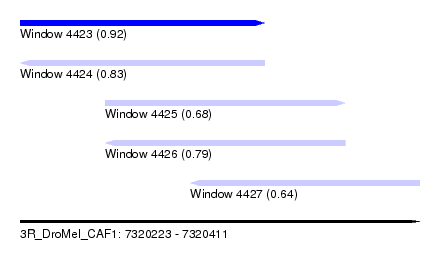
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,320,223 – 7,320,411 |
| Length | 188 |
| Max. P | 0.920659 |

| Location | 7,320,223 – 7,320,338 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.18 |
| Mean single sequence MFE | -38.18 |
| Consensus MFE | -33.97 |
| Energy contribution | -34.85 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7320223 115 + 27905053 GGUUAGUAUCAACAAUUGGAUAUCUGUGUUUGUCGGCACAGGGUUAAAUUGCCCAUUGAAAGGCCGCCGAGGGCAAACAAUCGUAGAUGCAUUUGCCGUAGUUGCAGUUGCAACU----- (((..(((((.((.((((....(((((((......)))))))......((((((.(((.........))))))))).)))).)).)))))....)))..((((((....))))))----- ( -37.40) >DroSim_CAF1 370 115 + 1 GGUUAGUAUCAACAAUUGGAUAUCUCUGUUUGUCGGCACUGGGUUAAAUUGCCUAUUGAAAGGCCGCCGAGGGCAAACAAUCGUAGAUGCAUUUGCCGUAGUUGCAGUUGCAACU----- (((..(((((.((.((((....((((.((..(((..((.(((((......))))).))...))).)).)))).....)))).)).)))))....)))..((((((....))))))----- ( -34.50) >DroEre_CAF1 386 115 + 1 GGUUAGUAUCAACAAUUGGAUAUCUGUGUUUGUCGGCACUGGGUUAAAUUGCCCAUUGAAAGGCCGCCGGGGGCAAACAAUCGUAGAUGCUUUUGCCGUAGUUGCAGUUGCAACU----- (((.((((((.((.((((......(((.((((.((((..(((((......))))).......)))).)))).)))..)))).)).))))))...)))..((((((....))))))----- ( -38.90) >DroYak_CAF1 386 120 + 1 GGUUAGUAUCAACAAUUGGAUAUCUGUGUUUGUCGGCACUGGCUUAAAUUGCCCAUUGAAAGGCCGCCGAGGGCAAACAAUCGUAGAUGCUUUUGCCGUAGUUGCAGUUGCAACUACCAA (((.((((((.((.((((......(((.((((.((((.(((((.......))).......)))))).)))).)))..)))).)).))))))...)))((((((((....))))))))... ( -41.91) >consensus GGUUAGUAUCAACAAUUGGAUAUCUGUGUUUGUCGGCACUGGGUUAAAUUGCCCAUUGAAAGGCCGCCGAGGGCAAACAAUCGUAGAUGCAUUUGCCGUAGUUGCAGUUGCAACU_____ (((.((((((.((.((((......(((.((((.((((..(((((......))))).......)))).)))).)))..)))).)).))))))...)))..((((((....))))))..... (-33.97 = -34.85 + 0.88)


| Location | 7,320,223 – 7,320,338 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.18 |
| Mean single sequence MFE | -31.13 |
| Consensus MFE | -27.41 |
| Energy contribution | -27.98 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7320223 115 - 27905053 -----AGUUGCAACUGCAACUACGGCAAAUGCAUCUACGAUUGUUUGCCCUCGGCGGCCUUUCAAUGGGCAAUUUAACCCUGUGCCGACAAACACAGAUAUCCAAUUGUUGAUACUAACC -----((((((....))))))..((((((((.(((...))))))))))).((((((((((......)))).........(((((........))))).........))))))........ ( -30.80) >DroSim_CAF1 370 115 - 1 -----AGUUGCAACUGCAACUACGGCAAAUGCAUCUACGAUUGUUUGCCCUCGGCGGCCUUUCAAUAGGCAAUUUAACCCAGUGCCGACAAACAGAGAUAUCCAAUUGUUGAUACUAACC -----((((((....)))))).((((((.((.((((....(((((((...(((((.((((......)))).(((......)))))))))))))))))))...)).))))))......... ( -30.90) >DroEre_CAF1 386 115 - 1 -----AGUUGCAACUGCAACUACGGCAAAAGCAUCUACGAUUGUUUGCCCCCGGCGGCCUUUCAAUGGGCAAUUUAACCCAGUGCCGACAAACACAGAUAUCCAAUUGUUGAUACUAACC -----((((((....)))))).(((((...........((..(..((((...))))..)..))..((((........)))).)))))........((.((((........)))))).... ( -27.80) >DroYak_CAF1 386 120 - 1 UUGGUAGUUGCAACUGCAACUACGGCAAAAGCAUCUACGAUUGUUUGCCCUCGGCGGCCUUUCAAUGGGCAAUUUAAGCCAGUGCCGACAAACACAGAUAUCCAAUUGUUGAUACUAACC ...((((((((....))))))))(((((..(((........))))))))...(((.((((......)))).......)))((((.((((((..............)))))).)))).... ( -35.04) >consensus _____AGUUGCAACUGCAACUACGGCAAAAGCAUCUACGAUUGUUUGCCCUCGGCGGCCUUUCAAUGGGCAAUUUAACCCAGUGCCGACAAACACAGAUAUCCAAUUGUUGAUACUAACC .....((((((....)))))).((((((.((.((((.....((((((...((((((((((......))))............)))))))))))).))))...)).))))))......... (-27.41 = -27.98 + 0.56)


| Location | 7,320,263 – 7,320,376 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.85 |
| Mean single sequence MFE | -40.00 |
| Consensus MFE | -32.47 |
| Energy contribution | -32.35 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7320263 113 + 27905053 GGGUUAAAUUGCCCAUUGAAAGGCCGCCGAGGGCAAACAAUCGUAGAUGCAUUUGCCGUAGUUGCAGUUGCAACU-------ACUAAGGCCAUCGAGUGCUGCAGUGUCAGCUCACCGAU ((((......)))).......((((......((((((..(((...)))...))))))((((((((....))))))-------))...))))((((.(((..((.......)).))))))) ( -41.80) >DroSim_CAF1 410 113 + 1 GGGUUAAAUUGCCUAUUGAAAGGCCGCCGAGGGCAAACAAUCGUAGAUGCAUUUGCCGUAGUUGCAGUUGCAACU-------ACCAAGGCCAUCGAGUGCUGCAGUGUCAGCUCACCGCU (((((..(((((...((((..((((......((((((..(((...)))...))))))((((((((....))))))-------))...)))).)))).....)))))...)))))...... ( -38.60) >DroEre_CAF1 426 113 + 1 GGGUUAAAUUGCCCAUUGAAAGGCCGCCGGGGGCAAACAAUCGUAGAUGCUUUUGCCGUAGUUGCAGUUGCAACU-------ACCAAGGCCAUCGAGUGCUGCAGUGUCAGCUCACCGAU ((((......)))).......((((......((((((..(((...)))...))))))((((((((....))))))-------))...))))((((.(((..((.......)).))))))) ( -41.40) >DroYak_CAF1 426 120 + 1 GGCUUAAAUUGCCCAUUGAAAGGCCGCCGAGGGCAAACAAUCGUAGAUGCUUUUGCCGUAGUUGCAGUUGCAACUACCAACUACCAAGGCCAACGAGUGCUGCAGUGUCAGCUCACCGAU ((((...(((((.((((....((((....((((((((..(((...)))...))))))((((((((....))))))))...)).....))))....))))..)))))...))))....... ( -38.20) >consensus GGGUUAAAUUGCCCAUUGAAAGGCCGCCGAGGGCAAACAAUCGUAGAUGCAUUUGCCGUAGUUGCAGUUGCAACU_______ACCAAGGCCAUCGAGUGCUGCAGUGUCAGCUCACCGAU ((((......)))).......((((....(.((((((..(((...)))...)))))).)((((((....))))))............)))).....(.((((......)))).)...... (-32.47 = -32.35 + -0.12)

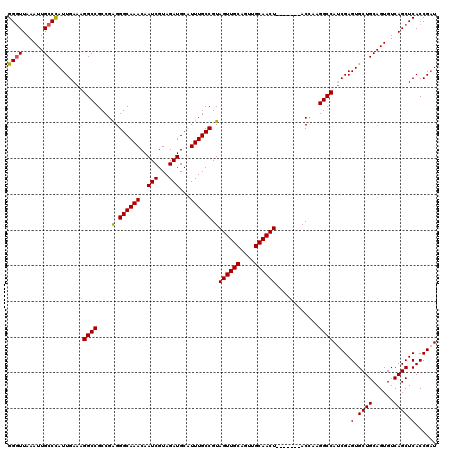
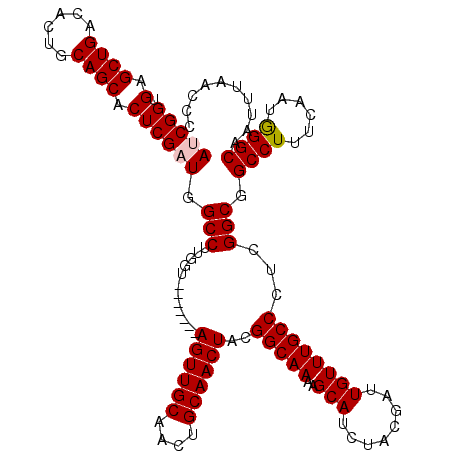
| Location | 7,320,263 – 7,320,376 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.85 |
| Mean single sequence MFE | -40.38 |
| Consensus MFE | -32.39 |
| Energy contribution | -32.70 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7320263 113 - 27905053 AUCGGUGAGCUGACACUGCAGCACUCGAUGGCCUUAGU-------AGUUGCAACUGCAACUACGGCAAAUGCAUCUACGAUUGUUUGCCCUCGGCGGCCUUUCAAUGGGCAAUUUAACCC (((((.(.((((......)))).)))))).(((...((-------((((((....))))))))((((((((.(((...)))))))))))...))).((((......)))).......... ( -40.60) >DroSim_CAF1 410 113 - 1 AGCGGUGAGCUGACACUGCAGCACUCGAUGGCCUUGGU-------AGUUGCAACUGCAACUACGGCAAAUGCAUCUACGAUUGUUUGCCCUCGGCGGCCUUUCAAUAGGCAAUUUAACCC .((((((......))))))...........(((...((-------((((((....))))))))((((((((.(((...)))))))))))...))).((((......)))).......... ( -40.20) >DroEre_CAF1 426 113 - 1 AUCGGUGAGCUGACACUGCAGCACUCGAUGGCCUUGGU-------AGUUGCAACUGCAACUACGGCAAAAGCAUCUACGAUUGUUUGCCCCCGGCGGCCUUUCAAUGGGCAAUUUAACCC (((((.(.((((......)))).)))))).(((...((-------((((((....))))))))(((((..(((........))))))))...))).((((......)))).......... ( -39.40) >DroYak_CAF1 426 120 - 1 AUCGGUGAGCUGACACUGCAGCACUCGUUGGCCUUGGUAGUUGGUAGUUGCAACUGCAACUACGGCAAAAGCAUCUACGAUUGUUUGCCCUCGGCGGCCUUUCAAUGGGCAAUUUAAGCC ...(((..((((......)))).((((((((....(((.((((((((((((....))))))))(((((..(((........))))))))..)))).)))..))))))))........))) ( -41.30) >consensus AUCGGUGAGCUGACACUGCAGCACUCGAUGGCCUUGGU_______AGUUGCAACUGCAACUACGGCAAAAGCAUCUACGAUUGUUUGCCCUCGGCGGCCUUUCAAUGGGCAAUUUAACCC (((((.(.((((......)))).)))))).(((............((((((....))))))..(((((..(((........))))))))...))).((((......)))).......... (-32.39 = -32.70 + 0.31)

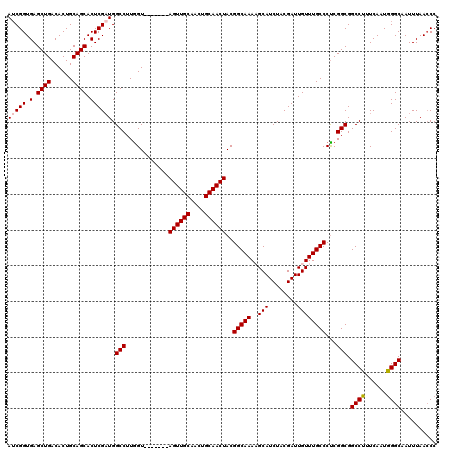
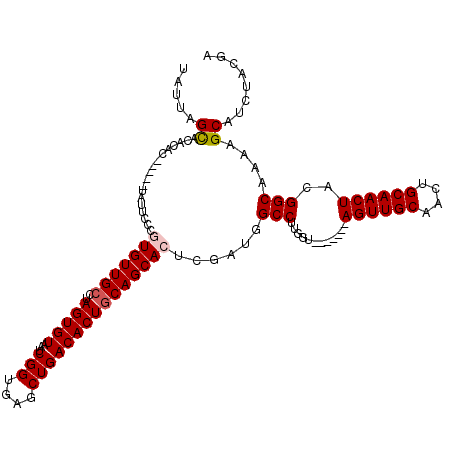
| Location | 7,320,303 – 7,320,411 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.16 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -23.07 |
| Energy contribution | -23.57 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7320303 108 - 27905053 UAUUAGCACACAC-----UUUUCCCUUGUUGCCUAGUGUAAUCGGUGAGCUGACACUGCAGCACUCGAUGGCCUUAGU-------AGUUGCAACUGCAACUACGGCAAAUGCAUCUACGA .....(((.....-----........((((((..((((...((((....)))))))))))))).......(((...((-------((((((....)))))))))))...)))........ ( -31.50) >DroSim_CAF1 450 108 - 1 UAUUAGCACACAC-----UUUUCCUUUGUUGGCUAGUGUAAGCGGUGAGCUGACACUGCAGCACUCGAUGGCCUUGGU-------AGUUGCAACUGCAACUACGGCAAAUGCAUCUACGA .....((..((((-----(...((......))..)))))..((((((......)))))).))....(((((((...((-------((((((....))))))))))).....))))..... ( -35.10) >DroEre_CAF1 466 113 - 1 UAUUAGUACACAUACUCGUAUUCCUGUGUUGCCUAGUGUAAUCGGUGAGCUGACACUGCAGCACUCGAUGGCCUUGGU-------AGUUGCAACUGCAACUACGGCAAAAGCAUCUACGA ...............(((((.....(((((((..((((...((((....)))))))))))))))..(((((((...((-------((((((....))))))))))).....))))))))) ( -36.50) >DroYak_CAF1 466 114 - 1 UAUUAGUGCACAU-----UAU-ACCGUGUUGCCUAGUGUAAUCGGUGAGCUGACACUGCAGCACUCGUUGGCCUUGGUAGUUGGUAGUUGCAACUGCAACUACGGCAAAAGCAUCUACGA ...((((((.((.-----..(-((((....(((.(((((...(((((......)))))..)))))....)))..)))))..))((((((((....)))))))).......))).)))... ( -36.90) >consensus UAUUAGCACACAC_____UAUUCCCGUGUUGCCUAGUGUAAUCGGUGAGCUGACACUGCAGCACUCGAUGGCCUUGGU_______AGUUGCAACUGCAACUACGGCAAAAGCAUCUACGA .....((..................(((((((..(((((...(((....)))))))))))))))......(((............((((((....))))))..)))....))........ (-23.07 = -23.57 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:17 2006