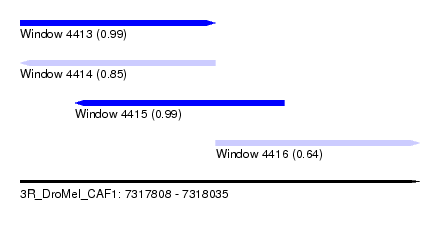
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,317,808 – 7,318,035 |
| Length | 227 |
| Max. P | 0.991739 |

| Location | 7,317,808 – 7,317,919 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.36 |
| Mean single sequence MFE | -44.82 |
| Consensus MFE | -43.74 |
| Energy contribution | -43.58 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7317808 111 + 27905053 ---------CGGAUACGGGUGCGGAUGUCGGGGCGGAGCCAUGACUACGCCAAAUGGCAAAACAAACUCGAACUGCGGCUCCGUUUCCGCAUUUCGAGGUUGAGUUGCGGCCCUGACGAA ---------((.((....)).))..(((((((((.(.(((((...........)))))....(((.((((((.(((((........))))).)))))).))).....).))))))))).. ( -44.00) >DroSec_CAF1 10682 111 + 1 ---------CGGAUACGGGUGCGGAUGUCGGGGCGGAGCCAUGACUACGCCAAAUGGCAAAACAAACUCGAACUGCGGCUCCGUUUCCGCAUUUCGAGGUUGAGUUGCGGCCCUGACGAA ---------((.((....)).))..(((((((((.(.(((((...........)))))....(((.((((((.(((((........))))).)))))).))).....).))))))))).. ( -44.00) >DroSim_CAF1 7713 111 + 1 ---------CGGAUACGGGUGCGGAUGUCGGGGCGGAGCCAUGACUACGCCAAAUGGCAAAACAAACUCGAACUGCGGCUCCGUUUCCGCAUUUCGAGGUUGAGUUGCGGCCCUGACGAA ---------((.((....)).))..(((((((((.(.(((((...........)))))....(((.((((((.(((((........))))).)))))).))).....).))))))))).. ( -44.00) >DroEre_CAF1 10770 117 + 1 ---CGGAUACGGAUACGGGUGCGGAUGUUGGGGCGGAGCCAUGACUACGCCAAAUGGCAAAACAAACUCGAACUGCGGCUCCGUUUCCGCAUUUCGAGGUUGAGUUGCGGCCCUGACGAA ---((.(..((....))..).))..(((..((((.(.(((((...........)))))....(((.((((((.(((((........))))).)))))).))).....).))))..))).. ( -44.90) >DroYak_CAF1 5259 120 + 1 AUGGGGAUGCGGAUACGGGUGCGGAUGUCGGGGCGGAGCCAUGACUACGCCAAAUGGCAAAACAAACUCGAACUGCGGCUCCGUUUCCGCAUUUCGAGGUUGAGUUGCGGCCCUGACGAA ....(.((.((....)).)).)...(((((((((.(.(((((...........)))))....(((.((((((.(((((........))))).)))))).))).....).))))))))).. ( -47.20) >consensus _________CGGAUACGGGUGCGGAUGUCGGGGCGGAGCCAUGACUACGCCAAAUGGCAAAACAAACUCGAACUGCGGCUCCGUUUCCGCAUUUCGAGGUUGAGUUGCGGCCCUGACGAA .........((.((....)).))..(((((((((.(.(((((...........)))))....(((.((((((.(((((........))))).)))))).))).....).))))))))).. (-43.74 = -43.58 + -0.16)



| Location | 7,317,808 – 7,317,919 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.36 |
| Mean single sequence MFE | -38.94 |
| Consensus MFE | -38.30 |
| Energy contribution | -38.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7317808 111 - 27905053 UUCGUCAGGGCCGCAACUCAACCUCGAAAUGCGGAAACGGAGCCGCAGUUCGAGUUUGUUUUGCCAUUUGGCGUAGUCAUGGCUCCGCCCCGACAUCCGCACCCGUAUCCG--------- ...(((.((...((((..(((.((((((.(((((........))))).)))))).)))..)))).....((((.(((....))).))))))))).................--------- ( -39.00) >DroSec_CAF1 10682 111 - 1 UUCGUCAGGGCCGCAACUCAACCUCGAAAUGCGGAAACGGAGCCGCAGUUCGAGUUUGUUUUGCCAUUUGGCGUAGUCAUGGCUCCGCCCCGACAUCCGCACCCGUAUCCG--------- ...(((.((...((((..(((.((((((.(((((........))))).)))))).)))..)))).....((((.(((....))).))))))))).................--------- ( -39.00) >DroSim_CAF1 7713 111 - 1 UUCGUCAGGGCCGCAACUCAACCUCGAAAUGCGGAAACGGAGCCGCAGUUCGAGUUUGUUUUGCCAUUUGGCGUAGUCAUGGCUCCGCCCCGACAUCCGCACCCGUAUCCG--------- ...(((.((...((((..(((.((((((.(((((........))))).)))))).)))..)))).....((((.(((....))).))))))))).................--------- ( -39.00) >DroEre_CAF1 10770 117 - 1 UUCGUCAGGGCCGCAACUCAACCUCGAAAUGCGGAAACGGAGCCGCAGUUCGAGUUUGUUUUGCCAUUUGGCGUAGUCAUGGCUCCGCCCCAACAUCCGCACCCGUAUCCGUAUCCG--- .......(((..((((..(((.((((((.(((((........))))).)))))).)))..)))).....((((.(((....))).))))............))).............--- ( -38.30) >DroYak_CAF1 5259 120 - 1 UUCGUCAGGGCCGCAACUCAACCUCGAAAUGCGGAAACGGAGCCGCAGUUCGAGUUUGUUUUGCCAUUUGGCGUAGUCAUGGCUCCGCCCCGACAUCCGCACCCGUAUCCGCAUCCCCAU ...(((.((...((((..(((.((((((.(((((........))))).)))))).)))..)))).....((((.(((....))).)))))))))....((..........))........ ( -39.40) >consensus UUCGUCAGGGCCGCAACUCAACCUCGAAAUGCGGAAACGGAGCCGCAGUUCGAGUUUGUUUUGCCAUUUGGCGUAGUCAUGGCUCCGCCCCGACAUCCGCACCCGUAUCCG_________ .......(((..((((..(((.((((((.(((((........))))).)))))).)))..)))).....((((.(((....))).))))............)))................ (-38.30 = -38.30 + 0.00)



| Location | 7,317,839 – 7,317,958 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.49 |
| Mean single sequence MFE | -44.56 |
| Consensus MFE | -44.20 |
| Energy contribution | -44.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7317839 119 - 27905053 ACUUUCC-CCGUCUUUUCCGACUGCAGGCAGGCGGUGCAAUUCGUCAGGGCCGCAACUCAACCUCGAAAUGCGGAAACGGAGCCGCAGUUCGAGUUUGUUUUGCCAUUUGGCGUAGUCAU .......-...........((((((.(((.(((((......)))))...)))((((..(((.((((((.(((((........))))).)))))).)))..))))........)))))).. ( -44.20) >DroSec_CAF1 10713 120 - 1 GCUUUCCACCGGCUUUUCCGACUGCAGGCAGGCGGUGCAAUUCGUCAGGGCCGCAACUCAACCUCGAAAUGCGGAAACGGAGCCGCAGUUCGAGUUUGUUUUGCCAUUUGGCGUAGUCAU (((.......)))......((((((.(((.(((((......)))))...)))((((..(((.((((((.(((((........))))).)))))).)))..))))........)))))).. ( -45.10) >DroSim_CAF1 7744 120 - 1 GCUUUCCACCGGCUUUUCCGACUGCAGGCAGGCGGUGCAAUUCGUCAGGGCCGCAACUCAACCUCGAAAUGCGGAAACGGAGCCGCAGUUCGAGUUUGUUUUGCCAUUUGGCGUAGUCAU (((.......)))......((((((.(((.(((((......)))))...)))((((..(((.((((((.(((((........))))).)))))).)))..))))........)))))).. ( -45.10) >DroEre_CAF1 10807 119 - 1 GCUUCUC-CCGUCUUUUCCGACUGCAGGCAGGCGGUGCAAUUCGUCAGGGCCGCAACUCAACCUCGAAAUGCGGAAACGGAGCCGCAGUUCGAGUUUGUUUUGCCAUUUGGCGUAGUCAU .......-...........((((((.(((.(((((......)))))...)))((((..(((.((((((.(((((........))))).)))))).)))..))))........)))))).. ( -44.20) >DroYak_CAF1 5299 119 - 1 GUUUUUC-CCGUCUUUUCCGACUGCAGGCAGGCGGUGCAAUUCGUCAGGGCCGCAACUCAACCUCGAAAUGCGGAAACGGAGCCGCAGUUCGAGUUUGUUUUGCCAUUUGGCGUAGUCAU .......-...........((((((.(((.(((((......)))))...)))((((..(((.((((((.(((((........))))).)))))).)))..))))........)))))).. ( -44.20) >consensus GCUUUCC_CCGUCUUUUCCGACUGCAGGCAGGCGGUGCAAUUCGUCAGGGCCGCAACUCAACCUCGAAAUGCGGAAACGGAGCCGCAGUUCGAGUUUGUUUUGCCAUUUGGCGUAGUCAU ...................((((((.(((.(((((......)))))...)))((((..(((.((((((.(((((........))))).)))))).)))..))))........)))))).. (-44.20 = -44.20 + -0.00)


| Location | 7,317,919 – 7,318,035 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -40.56 |
| Consensus MFE | -28.12 |
| Energy contribution | -28.25 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7317919 116 + 27905053 UUGCACCGCCUGCCUGCAGUCGGAAAAGACGG-GGAAAGUCCGGGA--AAUGCCUGGGAAAGGUCGGGGAAAAACCGGAAAGAGGGGUCGCGGCGGUACACGCGCGUUAAGUGGUGCAG .((((((((..((((...(((......)))..-......(((((..--..(.(((((......))))).)....))))).....))))((((.((.....))))))....)))))))). ( -42.80) >DroSec_CAF1 10793 106 + 1 UUGCACCGCCUGCCUGCAGUCGGAAAAGCCGGUGGAAAGCCCGGGA-------------AAGGCCGGGGAAAAACCGGAAAGAGGGGUCGCGGCGGUACACGCGCGUUAAGUGGUGCAG .(((((((((((((.((..((.......(((((......(((((..-------------....))))).....))))).......))..)))))))...((....))...)))))))). ( -42.04) >DroSim_CAF1 7824 105 + 1 UUGCACCGCCUGCCUGCAGUCGGAAAAGCCGGUGGAAAGCCCGGGA-------------AA-ACCGAGGAAAAACCGGAAAGUGGGGUCGCGGCGGUACACGCGCGUUAAGUGGUGCAG .((((((((...((..(..((((.....))(((......(((((..-------------..-.))).))....))).))..)..))..((((.((.....))))))....)))))))). ( -41.30) >DroYak_CAF1 5379 111 + 1 UUGCACCGCCUGCCUGCAGUCGGAAAAGACGG-GAAAAAC-AG---GAAGAGAC-GGGAAAGGCCG-GGAAAAUGUGGAA-AAGGGGACGCGGCGGUACACGCGCGUUAAGUGGUGCAG .((((((((((..(..(((((......)))..-.......-..---.......(-((......)))-......))..)..-.))..((((((.((.....))))))))..)))))))). ( -36.10) >consensus UUGCACCGCCUGCCUGCAGUCGGAAAAGACGG_GGAAAGCCCGGGA_____________AAGGCCGGGGAAAAACCGGAAAGAGGGGUCGCGGCGGUACACGCGCGUUAAGUGGUGCAG .((((((((...(((....(((.......)))...............................((((.......)))).....)))..((((.((.....))))))....)))))))). (-28.12 = -28.25 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:06 2006