
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,315,873 – 7,316,137 |
| Length | 264 |
| Max. P | 0.889050 |

| Location | 7,315,873 – 7,315,988 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.26 |
| Mean single sequence MFE | -40.00 |
| Consensus MFE | -26.37 |
| Energy contribution | -29.45 |
| Covariance contribution | 3.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7315873 115 + 27905053 UUUGUUGCCGGGGUACGUGAGACCCGGCCGGGCU--ACCACCCCC-CUGCC-GAUUCAACCCCCCUUCCUGUCGACGGCGGCGACAAAAGUCUUUAAUUUCUGGCCAAUACCAACGGCA- .....(((((((((.(....)))))(((((((..--.........-(((((-(..((.((..........)).)))))))).(((....))).......)))))))........)))))- ( -39.20) >DroSec_CAF1 8745 117 + 1 UUUGUUGCCCGGGUACGUGAGACCCGGCCGGGCUUCACCACCCCCGCCACU-GAUUCAACCC-CCUUCCUGUCGACGGCGGCGACAAAAGUCUUUAAUUUCUGGCCAAUACCAACGGCA- .....((((.((((.(....)))))(((((((...........(((((..(-(((.......-.......))))..))))).(((....))).......))))))).........))))- ( -38.64) >DroSim_CAF1 4158 113 + 1 UUUGUUGCCCGGGUACGUGAGACCCGGCCCGGCUCCUCCACCGCC----CU-GAUUCAACCC-CCAUCCUGUCGACGGCGGCGACAAAAGUCUUUAAUUUCUGGCCAAUACCAACGGCA- .((((((.((((((.(....)))))))...((((......(((((----.(-(((.......-.......))))..))))).(((....)))..........)))).....))))))..- ( -34.24) >DroEre_CAF1 8690 104 + 1 UUUGUUGCCGGGGUACGUGAGACCCAGCCGGGCUC---------------GCCCCAAAACCC-CCUUUCUCUUGACGGCGGCGACGAAAGUCUUUAAUUUCUGGCCAAUACCAACGGCAA ....((((((((((.(....))))).(((((((.(---------------(((.(((.....-........)))..))))))(((....)))........))))).........)))))) ( -39.62) >DroYak_CAF1 3280 107 + 1 UUUGUUGCCGGGGUACGUGAGACCCAGCCGGGCAU------------UGGGGGGUUAAACCC-CCUUCCUGUCGACGGCGGCGACAAAAGUCUUUAAUUUCUGGCCAAUACCAACGGCAA (((((((((.((((.(....))))).((((((((.------------.((((((.....)))-)))...))))..)))))))))))))...............(((.........))).. ( -48.30) >consensus UUUGUUGCCGGGGUACGUGAGACCCGGCCGGGCUC__CCACC_CC____CG_GAUUCAACCC_CCUUCCUGUCGACGGCGGCGACAAAAGUCUUUAAUUUCUGGCCAAUACCAACGGCA_ (((((((((.((((.(....))))).(((((((.....................................)))..)))))))))))))...............(((.........))).. (-26.37 = -29.45 + 3.08)

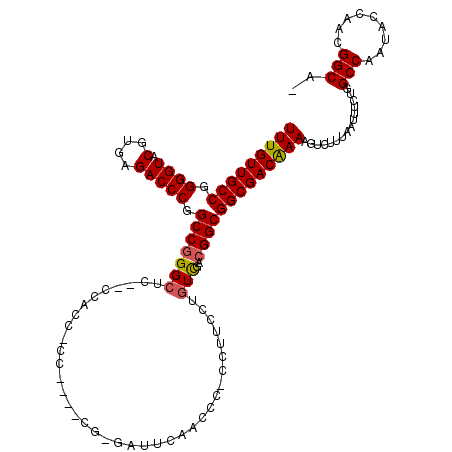

| Location | 7,315,949 – 7,316,057 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.24 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -26.56 |
| Energy contribution | -26.56 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.710848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7315949 108 + 27905053 GCGACAAAAGUCUUUAAUUUCUGGCCAAUACCAACGGCA------------GCAGCUACGGCAAUGCCGACGAUGCCAAAAGCUUUUCAAAGCCACCGAAACGGGGGCGUUAACAAGCGC (((.(....(((.........(((......)))..((((------------...((....))..)))))))((((((....((((....))))..(((...))).)))))).....)))) ( -30.90) >DroSec_CAF1 8823 108 + 1 GCGACAAAAGUCUUUAAUUUCUGGCCAAUACCAACGGCA------------ACAGCUACGGCAAUGUCGACGAUGCCAAAAGCUUUUCAAAGCCACCGAAACGGGGGCGUUAACAAGCGC (((.(....(((.........(((......)))..((((------------...((....))..)))))))((((((....((((....))))..(((...))).)))))).....)))) ( -27.10) >DroSim_CAF1 4232 108 + 1 GCGACAAAAGUCUUUAAUUUCUGGCCAAUACCAACGGCA------------ACAGCUACGGCAAUGCCGACGAUGCCAAAAGCUUUUCAAAGCCACCGAAACGGGGGCGUUAACAAGCGC (((.(....(((.........(((......)))..((((------------...((....))..)))))))((((((....((((....))))..(((...))).)))))).....)))) ( -29.80) >DroEre_CAF1 8754 120 + 1 GCGACGAAAGUCUUUAAUUUCUGGCCAAUACCAACGGCAAUGGCUACGGCAAUGGCUACGGCAAUGCCAACGAUGCCAAAAGCUUUUCAAAGCCACCGAAACGGGGGCGUUAACAAGCGC (((((....))).........((((((...((...))...)))))).((((...((....))..))))...((((((....((((....))))..(((...))).)))))).....)).. ( -37.50) >DroYak_CAF1 3347 120 + 1 GCGACAAAAGUCUUUAAUUUCUGGCCAAUACCAACGGCAACGAGAACGGCAACAGCUACGGCAAUGCCAACGAUGCCAAAAGCUUUUCAAAGCCACCGAAACGGGGGCGUUAACAAGCGC (((((....))).........((((.........((....)).....((((...((....))..))))......))))...((((......(((.(((...))).)))......)))))) ( -33.30) >consensus GCGACAAAAGUCUUUAAUUUCUGGCCAAUACCAACGGCA____________ACAGCUACGGCAAUGCCGACGAUGCCAAAAGCUUUUCAAAGCCACCGAAACGGGGGCGUUAACAAGCGC (((((....)))...........(((.........)))................))...((((.((....)).))))....((((......(((.(((...))).)))......)))).. (-26.56 = -26.56 + 0.00)



| Location | 7,316,017 – 7,316,137 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -31.82 |
| Energy contribution | -32.42 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7316017 120 - 27905053 UCUUUUUUUCCAGUGCAUCUGGCUGUGUGUCUUUAUCUAGCUGUGUUGUAGUUGGAAAUAAACGGCAAGUUUUGUCUCUGGCGCUUGUUAACGCCCCCGUUUCGGUGGCUUUGAAAAGCU .........((((.(((.((.(((((.(((.....((((((((.....)))))))).))).))))).))...)))..)))).((((.((((.(((.(((...))).))).)))).)))). ( -34.90) >DroSec_CAF1 8891 118 - 1 A-UUU-UUUCCAGUGCAUCUGGCUGUGUGUCUUUAUCUAGCUGUGUUGUAGUUGGAAAUAAACGGCAAGUUUUGUCUCUGGCGCUUGUUAACGCCCCCGUUUCGGUGGCUUUGAAAAGCU .-...-...((((.(((.((.(((((.(((.....((((((((.....)))))))).))).))))).))...)))..)))).((((.((((.(((.(((...))).))).)))).)))). ( -34.90) >DroSim_CAF1 4300 119 - 1 A-UUUUUUUCCAGUGCAUCUGGCUGUGUGUCUUUAUCUAGCUGUGUUGUAGUUGGAAAUAAACGGCAAGUUUUGUCUCUGGCGCUUGUUAACGCCCCCGUUUCGGUGGCUUUGAAAAGCU .-.......((((.(((.((.(((((.(((.....((((((((.....)))))))).))).))))).))...)))..)))).((((.((((.(((.(((...))).))).)))).)))). ( -34.90) >DroEre_CAF1 8834 118 - 1 A-UUU-UUGCCAGUGCAUCUGUCUGUGUGUCUUUAUCUAGCCGUGUUGUAGUUGGAAAUAAACGGCAAGUUUUGUCUCUGGCGCUUGUUAACGCCCCCGUUUCGGUGGCUUUGAAAAGCU .-...-..(((((.((((......))))...........(((((.((((........)))))))))...........)))))((((.((((.(((.(((...))).))).)))).)))). ( -32.70) >DroYak_CAF1 3427 118 - 1 G-UUU-UUUCCAGUGCAUCUGGCUGUGUGGCUUUAUCUAGCCGUGUUGUAGUUGGAAAUAAACGGCAAGUUUUGUCUCUGGCGCUUGUUAACGCCCCCGUUUCGGUGGCUUUGAAAAGCU (-(((-(((((((((((.(((((((.(((....))).)))))).).)))).))))))).))))((((.....))))......((((.((((.(((.(((...))).))).)))).)))). ( -36.60) >consensus A_UUU_UUUCCAGUGCAUCUGGCUGUGUGUCUUUAUCUAGCUGUGUUGUAGUUGGAAAUAAACGGCAAGUUUUGUCUCUGGCGCUUGUUAACGCCCCCGUUUCGGUGGCUUUGAAAAGCU .........((((.(((.((.(((((.........((((((((.....)))))))).....))))).))...)))..)))).((((.((((.(((.(((...))).))).)))).)))). (-31.82 = -32.42 + 0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:03 2006