| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,066,119 – 1,066,221 |
| Length | 102 |
| Max. P | 0.804657 |

| Location | 1,066,119 – 1,066,221 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
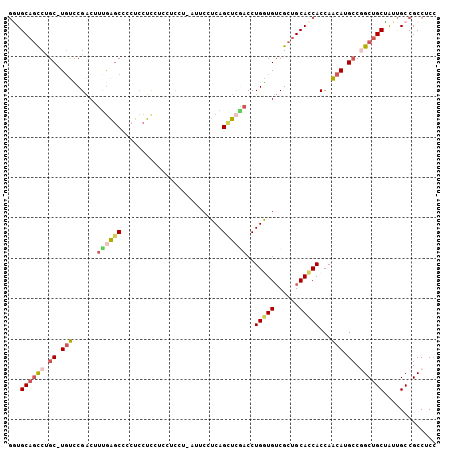
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -30.71 |
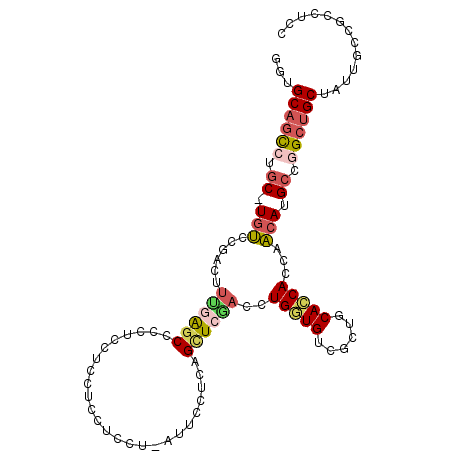
| Consensus MFE | -19.35 |
| Energy contribution | -20.16 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
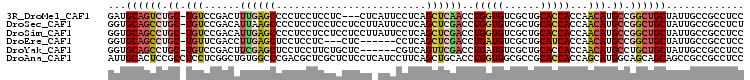
>3R_DroMel_CAF1 1066119 102 + 27905053 GAUGCAGUCUGC-UGUCCGACUUUGAGCCCCUCCUCCUC---CUCAUUCCUCAGCUCAACCUGGUGUCGCUGCACCACCAACAUGCCGGCUGCUAUUGCCGCCUCC ...((((((.((-(((......((((((...........---...........))))))..((((((....))))))...))).)).))))))............. ( -25.15) >DroSec_CAF1 2362 105 + 1 GGUGCAGCCUGC-UGUCCGACAUUAAGCCCCUCCUCCUCCUCCUUAUUCCUCAGCUCGACCUGGUGUCGCUGCACCACCAACAUGCCGGCUGCUAUUGCCGCCUCU ((((((((..((-((...((...((((...............)))).))..))))..(((.....)))))))))))...........(((.((....)).)))... ( -30.46) >DroSim_CAF1 2462 105 + 1 GGUGCAGCCUGC-UGUCCGACAUUGAGCCCCUCCUCCUCCUCCUUAUUCCUCAGCUCGACCUGGUGUCGCUGCACCACCAACAUGCCGGCUGCUAUUGCCGCCUCC ((((((((..((-...(((...((((((.........................))))))..))).)).))))))))...........(((.((....)).)))... ( -31.21) >DroEre_CAF1 2763 96 + 1 GGUGCAGCCUGC-UGUUCGACCUUGAGCUCCUCCUC---CUC------CCUCAGCUCGACCUGAUGUCGCUGCAUCACCAACAUGCCGGCUGCUAUUGCCGCCUCC (((((((((.((-((((.(...(((((((.......---...------....)))))))..((((((....))))))).)))).)).))))))....)))...... ( -30.84) >DroYak_CAF1 6041 99 + 1 GGUGCAGCCUGC-UGUCCGACUUCGAGCUCCUCCUUCUGCUC------CGUCAGUUCGACCUGAUGUCGCUGCACCACCAACAUGCCUGCUGCUAUUGCCGCCUCC ((((((((....-...........((((..........))))------((((((......))))))..))))))))............((.((....)).)).... ( -30.80) >DroAna_CAF1 1926 106 + 1 AUUGCACUCCGCCUCCUCGGCUGUGGCCCGACGCUCGCUCUCCUCAUCCUUCAGCUGCACCUGGUGGCGCCGCACCACCAGCAUGGCAGCAGCAGCCGCCGCCUCC ...((.....))......(((.(((((.((.....))................(((((..(((((((.......)))))))....)))))....))))).)))... ( -35.80) >consensus GGUGCAGCCUGC_UGUCCGACUUUGAGCCCCUCCUCCUCCUCCU_AUUCCUCAGCUCGACCUGGUGUCGCUGCACCACCAACAUGCCGGCUGCUAUUGCCGCCUCC ...((((((.((.(((......((((((.........................))))))..(((((......)))))...))).)).))))))............. (-19.35 = -20.16 + 0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:11 2006