
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,289,302 – 7,289,392 |
| Length | 90 |
| Max. P | 0.790094 |

| Location | 7,289,302 – 7,289,392 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.60 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -14.50 |
| Energy contribution | -14.42 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7289302 90 + 27905053 AAUGGUAUUUCUUU-GGCGCUGUUUGCCGGAAUAUCUGAUAUCCCCAAUGGUCCGAGUAGGCAGCGCACAAAAAAU-------------------AGAAGUCAACAAAAA ..((..((((((((-((((((((((((((((.(((.((.......))))).)))).))))))))))).))).....-------------------))))))...)).... ( -29.00) >DroSec_CAF1 10421 90 + 1 AAUGGUAUUUCUUU-GGCGCUGUUUGCCGGAAUAUCUGAUAUCCCCAAUGGUCCGAGUAGGCAGCGAACAAAUAAU-------------------AGAAGUCAACAAAAA ..((..((((((((-(.((((((((((((((.(((.((.......))))).)))).))))))))))..))).....-------------------))))))...)).... ( -24.40) >DroSim_CAF1 15010 90 + 1 AAUGGUAUUUCUUU-GGCGCUGUUUGCCGGAAUAUCUGAUAUCCCCAAUGGUCCGAGUAGGCAGCGAACAAAAAAU-------------------AGAAGUCAACAAAAA ..((..((((((((-(.((((((((((((((.(((.((.......))))).)))).))))))))))..))).....-------------------))))))...)).... ( -24.40) >DroEre_CAF1 10545 87 + 1 AAUGGUAUUUCUUU-GGCGCUGUUUGCCCGAAUAUCUGUUAUC--CGAUGGACGUAGUAGGCAGCUCACAAA-AAU-------------------AUAAGUCAACAAGAA ....((((((..((-((.(((((((((.((..((((.......--.))))..))..))))))))).).))))-)))-------------------))............. ( -18.90) >DroYak_CAF1 15748 88 + 1 AAUGGUAUUUCUCU-GGCGCUGUUUGCCCGAAUAUCUGAUAUC--CAAUGGUCGGAGUAGGCAGCUUACAAAAAAU-------------------AAAAGUCAACAAGAG .........(((.(-((((((((((((((((.(((.((.....--))))).)))).)))))))))(((.......)-------------------))..))))...))). ( -20.40) >DroAna_CAF1 10328 103 + 1 AAUGGUAUUCGUUUCGGGGCAGUUUGUCUGAAUUUCCA-------AAACGGUCAGAGUAGGCAGCACACAAAAAACGGGAACCUAACACGAACAGAACAGUCAACAGAAA ...(((.(((((((..(.((.((((((((((....((.-------....))))))).))))).))...)...))))))).)))........................... ( -19.60) >consensus AAUGGUAUUUCUUU_GGCGCUGUUUGCCGGAAUAUCUGAUAUC__CAAUGGUCCGAGUAGGCAGCGCACAAAAAAU___________________AGAAGUCAACAAAAA ..................(((((((((((((.(((............))).)))).)))))))))............................................. (-14.50 = -14.42 + -0.08)

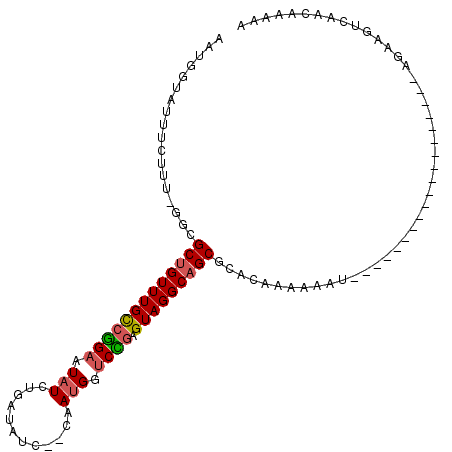
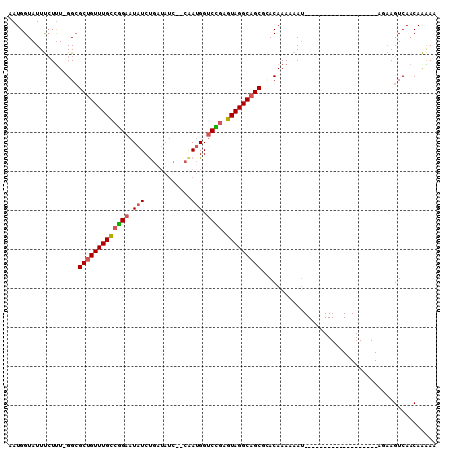
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:53 2006