
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,278,278 – 7,278,397 |
| Length | 119 |
| Max. P | 0.770488 |

| Location | 7,278,278 – 7,278,397 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.68 |
| Mean single sequence MFE | -41.97 |
| Consensus MFE | -27.26 |
| Energy contribution | -28.71 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
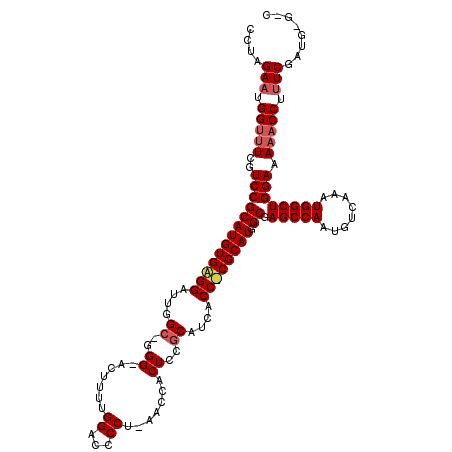
>3R_DroMel_CAF1 7278278 119 + 27905053 CCCAGAAUGGUUUCGUCCGCAUGUGAGGAUUGGC-GGGUACUUUUGGACCCCAAAACCACUACGCAUCACCUCGCAUGGCGAGCCAAUGUCAAAUGGCUGGAAAAACCUUUCGAUGCGCC ....(((.(((((..(((((((((((((....((-((((..((((((...))))))..))).)))....)))))))).)).(((((........)))))))).))))).)))........ ( -42.40) >DroSim_CAF1 3534 111 + 1 CCUAGAAUGGUUUCGUCCGCAUGUGGGGAUUGGCUGGGGACUUUUGGACCCCU-AACCACUCCGCAUCCCCCCGCAUGGCGAGCCAAUGUCAAAUGGCUGGAAAAACCUUUC-------- ....(((.(((((..(((((((((((((...((.((.(((....(((......-..))).))).)).)))))))))).)).(((((........)))))))).))))).)))-------- ( -41.00) >DroYak_CAF1 4536 109 + 1 CCUAGAUUGGC-UCGUCCGCAUGUGAGGAUUGGG--GG---UAUUGGACCCCU-A---ACUCCGCAUAACC-CGCAUGGCGAGCCAAUGUCAAAUGGCUGGAAAAACCUUUCGAUGUGGC .....((((((-(((((.((.((((.((((((((--((---(.....))))))-)---).))).))))...-.))..)))))))))))((((..(((..((.....))..)))...)))) ( -42.50) >consensus CCUAGAAUGGUUUCGUCCGCAUGUGAGGAUUGGC_GGG_ACUUUUGGACCCCU_AACCACUCCGCAUCACC_CGCAUGGCGAGCCAAUGUCAAAUGGCUGGAAAAACCUUUCGAUG_G_C ....(((.(((((..(((((((((((((....((..((.......((...)).......))..))....)))))))).)).(((((........)))))))).))))).)))........ (-27.26 = -28.71 + 1.45)


| Location | 7,278,278 – 7,278,397 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.68 |
| Mean single sequence MFE | -41.43 |
| Consensus MFE | -27.81 |
| Energy contribution | -29.70 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
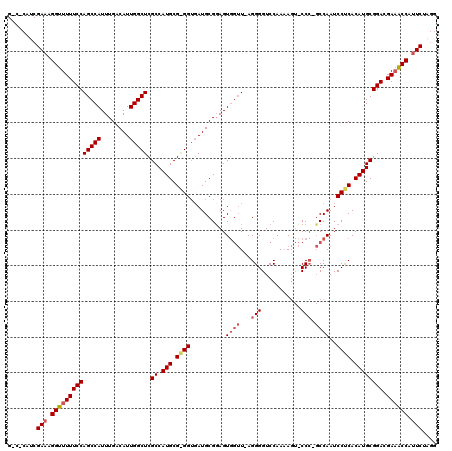
>3R_DroMel_CAF1 7278278 119 - 27905053 GGCGCAUCGAAAGGUUUUUCCAGCCAUUUGACAUUGGCUCGCCAUGCGAGGUGAUGCGUAGUGGUUUUGGGGUCCAAAAGUACCC-GCCAAUCCUCACAUGCGGACGAAACCAUUCUGGG ........(((.((((((((((((((........))))).((.(((.((((....(((..(((.((((((...)))))).))).)-))....)))).)))))))).)))))).))).... ( -41.30) >DroSim_CAF1 3534 111 - 1 --------GAAAGGUUUUUCCAGCCAUUUGACAUUGGCUCGCCAUGCGGGGGGAUGCGGAGUGGUU-AGGGGUCCAAAAGUCCCCAGCCAAUCCCCACAUGCGGACGAAACCAUUCUAGG --------(((.((((((((((((((........))))).((.(((.(((((.........(((((-.((((.(.....).))))))))).))))).)))))))).)))))).))).... ( -45.00) >DroYak_CAF1 4536 109 - 1 GCCACAUCGAAAGGUUUUUCCAGCCAUUUGACAUUGGCUCGCCAUGCG-GGUUAUGCGGAGU---U-AGGGGUCCAAUA---CC--CCCAAUCCUCACAUGCGGACGA-GCCAAUCUAGG .((...(((((.((((.....)))).))))).(((((((((((..(((-.....)))(((..---.-.(((((.....)---))--))...)))........)).)))-))))))...)) ( -38.00) >consensus G_C_CAUCGAAAGGUUUUUCCAGCCAUUUGACAUUGGCUCGCCAUGCG_GGUGAUGCGGAGUGGUU_AGGGGUCCAAAAGU_CCC_GCCAAUCCUCACAUGCGGACGAAACCAUUCUAGG ........(((.((((((((((((((........))))).((.(((.((((..........((((...(((...........))).))))..)))).)))))))).)))))).))).... (-27.81 = -29.70 + 1.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:49 2006