
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,276,403 – 7,276,538 |
| Length | 135 |
| Max. P | 0.793285 |

| Location | 7,276,403 – 7,276,513 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.38 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -23.30 |
| Energy contribution | -23.63 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7276403 110 + 27905053 AAAGUGCUUAAAAAAAAAACAAAUAUAUCGGACGGGAUAUUUUGGGAACUGGCCAUUCAUAACGCCCCUCCAGAGAGAUUUAUUAGAUCGAUUGGCCAGUGACAAGAGCU---------- .....((((..........((((.(((((......)))))))))(..((((((((.((.........((....)).((((.....)))))).))))))))..)..)))).---------- ( -29.30) >DroSim_CAF1 1492 102 + 1 AAAGUGCUUAAAAAAA--------AUAUCGGACGGGAUAUUUUGGGAACAGGCCAUUCAUAACGCCCCUCCAGAGAGAUUUAUUAGAUCGAUUGGCCAGUGACAAGAGCU---------- .....((((....(((--------(((((......)))))))).(..((.(((((.((.........((....)).((((.....)))))).))))).))..)..)))).---------- ( -24.40) >DroYak_CAF1 2066 108 + 1 AAAGUGCCUCAAA-----------AUAUCGGACG-GAUAUUUUGGGAACUGGCCAUUCAUAACGCCCCUCCAGAGAGAUUUAUUAGAUCGAUUGGCCAGUGACAAGAGCUGAAGCAAAGC ....(((((((((-----------(((((.....-))))))))))).((((((((.((.........((....)).((((.....)))))).)))))))).............))).... ( -32.50) >consensus AAAGUGCUUAAAAAAA________AUAUCGGACGGGAUAUUUUGGGAACUGGCCAUUCAUAACGCCCCUCCAGAGAGAUUUAUUAGAUCGAUUGGCCAGUGACAAGAGCU__________ .....((..................((((......))))(((((...((((((((.((.........((....)).((((.....)))))).))))))))..)))))))........... (-23.30 = -23.63 + 0.33)

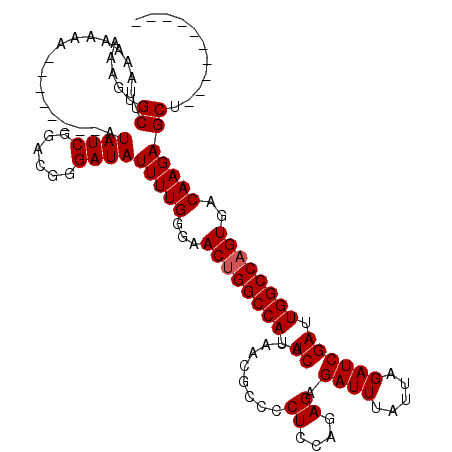

| Location | 7,276,403 – 7,276,513 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.38 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -22.62 |
| Energy contribution | -22.73 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7276403 110 - 27905053 ----------AGCUCUUGUCACUGGCCAAUCGAUCUAAUAAAUCUCUCUGGAGGGGCGUUAUGAAUGGCCAGUUCCCAAAAUAUCCCGUCCGAUAUAUUUGUUUUUUUUUUAAGCACUUU ----------.(((......((((((((.(((...((((...((((....))))...))))))).))))))))...(((((((((......))))).))))...........)))..... ( -25.80) >DroSim_CAF1 1492 102 - 1 ----------AGCUCUUGUCACUGGCCAAUCGAUCUAAUAAAUCUCUCUGGAGGGGCGUUAUGAAUGGCCUGUUCCCAAAAUAUCCCGUCCGAUAU--------UUUUUUUAAGCACUUU ----------.(((......((.(((((.(((...((((...((((....))))...))))))).))))).))....((((((((......)))))--------))).....)))..... ( -21.90) >DroYak_CAF1 2066 108 - 1 GCUUUGCUUCAGCUCUUGUCACUGGCCAAUCGAUCUAAUAAAUCUCUCUGGAGGGGCGUUAUGAAUGGCCAGUUCCCAAAAUAUC-CGUCCGAUAU-----------UUUGAGGCACUUU ....((((((..........((((((((.(((...((((...((((....))))...))))))).))))))))....((((((((-.....)))))-----------))))))))).... ( -32.10) >consensus __________AGCUCUUGUCACUGGCCAAUCGAUCUAAUAAAUCUCUCUGGAGGGGCGUUAUGAAUGGCCAGUUCCCAAAAUAUCCCGUCCGAUAU________UUUUUUUAAGCACUUU ...........(((.(((..((((((((.(((...((((...((((....))))...))))))).))))))))...))).(((((......)))))................)))..... (-22.62 = -22.73 + 0.11)

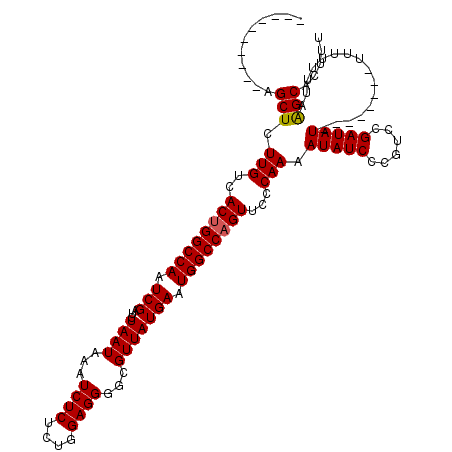

| Location | 7,276,443 – 7,276,538 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -28.98 |
| Energy contribution | -29.09 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7276443 95 - 27905053 UGUACAUUUGGGCGGCAAUAUGCGC-------------------------AGCUCUUGUCACUGGCCAAUCGAUCUAAUAAAUCUCUCUGGAGGGGCGUUAUGAAUGGCCAGUUCCCAAA ......((((((.(((((...((..-------------------------.))..)))))((((((((.(((...((((...((((....))))...))))))).)))))))).)))))) ( -30.40) >DroSim_CAF1 1524 95 - 1 UGUACAUUUGGGCGGCGAAAUGCGC-------------------------AGCUCUUGUCACUGGCCAAUCGAUCUAAUAAAUCUCUCUGGAGGGGCGUUAUGAAUGGCCUGUUCCCAAA .(((((...((((.(((.....)))-------------------------.)))).))).)).(((((.(((...((((...((((....))))...))))))).))))).......... ( -26.00) >DroYak_CAF1 2094 120 - 1 UGUACAUUUGGGCGGCGAAAUGCGCAGAGCUUGCGAAAGCGCUUUGCUUCAGCUCUUGUCACUGGCCAAUCGAUCUAAUAAAUCUCUCUGGAGGGGCGUUAUGAAUGGCCAGUUCCCAAA ......((((((.(((((...(((((((((..((....)))))))))....))..)))))((((((((.(((...((((...((((....))))...))))))).)))))))).)))))) ( -45.30) >consensus UGUACAUUUGGGCGGCGAAAUGCGC_________________________AGCUCUUGUCACUGGCCAAUCGAUCUAAUAAAUCUCUCUGGAGGGGCGUUAUGAAUGGCCAGUUCCCAAA ......((((((.(((((...((............................))..)))))((((((((.(((...((((...((((....))))...))))))).)))))))).)))))) (-28.98 = -29.09 + 0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:44 2006