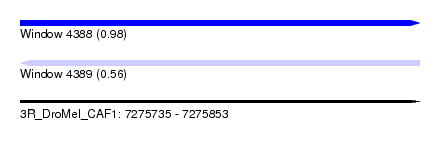
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,275,735 – 7,275,853 |
| Length | 118 |
| Max. P | 0.978560 |

| Location | 7,275,735 – 7,275,853 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.47 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -23.59 |
| Energy contribution | -23.70 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.92 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
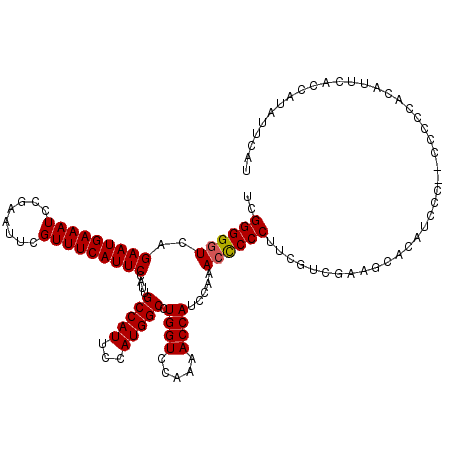
>3R_DroMel_CAF1 7275735 118 + 27905053 UCGGGGGUCAGAAUGAAAUCCGAAUUCGUUUCAUUCCAUUGCCAUUCCAUGGCCUGGUCCAAAACCAUCCA-ACCCCCUUCGUCGAAGCCCAUCCCU-CCCCCACAUUCACCAUAUUCAU ..((((((..(((((((((........)))))))))....(((((...))))).((((.....))))....-))))))...................-...................... ( -26.30) >DroSim_CAF1 822 120 + 1 UCGGGGGUCAGAAUGAAAUCCGAAUUCGUUUCAUUCCAUUGCCAUUCCAUGGCCUGGUCCAAAACCAUCCAAACCCCCUUCGGCGAAGCACAUCCCAACCCCCACAUCCACCAUACUCAU ..((((((..(((((((((........)))))))))..(((((......(((..((((.....)))).)))..........)))))...........))))))................. ( -29.19) >DroYak_CAF1 1429 112 + 1 UCGGGGGUCAGAAUGAAAUCCGAAUUCGUUUCAUUCCAUUGCCAUUCCAUGGCCUGGUCCAAAACCAGCCAAAAUCCCCUCCUC----CACAUCCU----CCACCAUUCCCCAUAUUCAU ..((((((..(((((((((........))))))))).............((((.((((.....))))))))..)))))).....----........----.................... ( -27.10) >consensus UCGGGGGUCAGAAUGAAAUCCGAAUUCGUUUCAUUCCAUUGCCAUUCCAUGGCCUGGUCCAAAACCAUCCAAACCCCCUUCGUCGAAGCACAUCCC__CCCCCACAUUCACCAUAUUCAU ..((((((..(((((((((........)))))))))....(((((...))))).((((.....)))).....)))))).......................................... (-23.59 = -23.70 + 0.11)


| Location | 7,275,735 – 7,275,853 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.47 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -26.40 |
| Energy contribution | -27.40 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
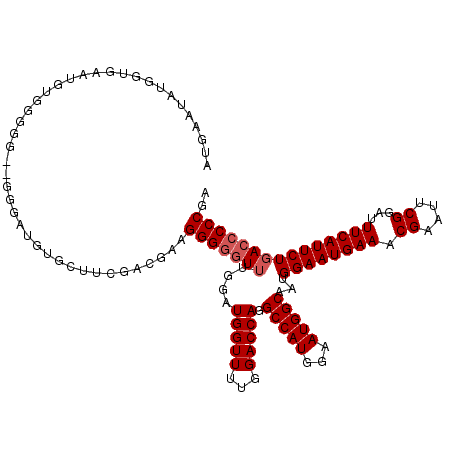
>3R_DroMel_CAF1 7275735 118 - 27905053 AUGAAUAUGGUGAAUGUGGGGG-AGGGAUGGGCUUCGACGAAGGGGGU-UGGAUGGUUUUGGACCAGGCCAUGGAAUGGCAAUGGAAUGAAACGAAUUCGGAUUUCAUUCUGACCCCCGA ................((((((-.((((((((.((((((....)....-......((((((..(((.(((((...)))))..)))..))))))....))))).))))))))..)))))). ( -37.60) >DroSim_CAF1 822 120 - 1 AUGAGUAUGGUGGAUGUGGGGGUUGGGAUGUGCUUCGCCGAAGGGGGUUUGGAUGGUUUUGGACCAGGCCAUGGAAUGGCAAUGGAAUGAAACGAAUUCGGAUUUCAUUCUGACCCCCGA ................(((((((..((((...((((...))))(..((((((((.((((((..(((.(((((...)))))..)))..))))))..))))))))..)))))..))))))). ( -44.60) >DroYak_CAF1 1429 112 - 1 AUGAAUAUGGGGAAUGGUGG----AGGAUGUG----GAGGAGGGGAUUUUGGCUGGUUUUGGACCAGGCCAUGGAAUGGCAAUGGAAUGAAACGAAUUCGGAUUUCAUUCUGACCCCCGA .......(((((..((((..----(((((((.----((((......)))).))..)))))..)))).(((((...)))))...((((((((.((....))...))))))))...))))). ( -29.50) >consensus AUGAAUAUGGUGAAUGUGGGGG__GGGAUGUGCUUCGACGAAGGGGGUUUGGAUGGUUUUGGACCAGGCCAUGGAAUGGCAAUGGAAUGAAACGAAUUCGGAUUUCAUUCUGACCCCCGA ..........................................(((((((....(((((...))))).(((((...)))))...((((((((.((....))...))))))))))))))).. (-26.40 = -27.40 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:41 2006