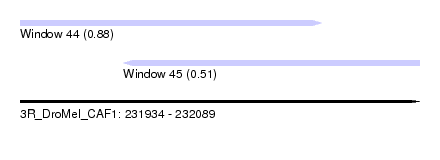
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 231,934 – 232,089 |
| Length | 155 |
| Max. P | 0.876311 |

| Location | 231,934 – 232,051 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -45.92 |
| Consensus MFE | -30.58 |
| Energy contribution | -31.42 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876311 |
| Prediction | RNA |
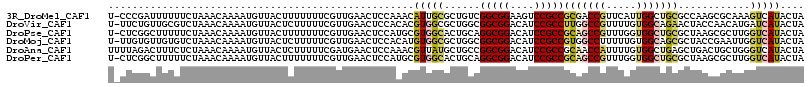
Download alignment: ClustalW | MAF
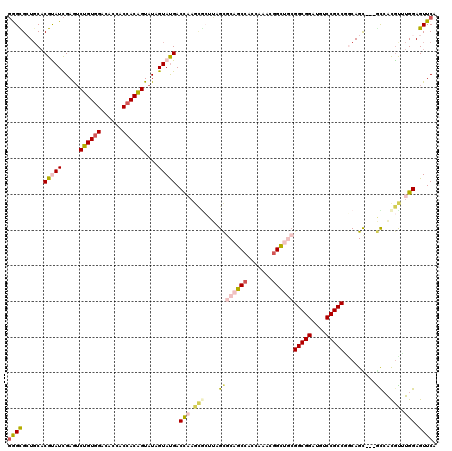
>3R_DroMel_CAF1 231934 117 + 27905053 GGGUGGUGCACGCAUCGAAGCUGUCGACACCACCACAGUAUAGUAUGACUUUGCGCUUGGCGCAGCCAAUGAACGGUCGCGGCGGACUUCCGCCGACAGC---GCAAUGUUUGGAGUUCA .((((((((((((......)).)).).)))))))............(((.(((((((.(..((.(((.......))).))(((((....)))))..))))---)))).)))......... ( -43.70) >DroPse_CAF1 1808 117 + 1 GAGCGCUGCACGUAUCGAGUCUGUGGACACCACCACAGUAUAGUAUGACCAAGCGCUUAGCGCAGCCACCAAACGGCUGCGGCGGAUGUCCGCCUGCAGU---GCCACGCAUGGAGUUCA ((((.((((.(((((.....((((((......))))))....))))).....(((((..((((((((.......))))))(((((....))))).)))))---))...))).)..)))). ( -49.20) >DroWil_CAF1 2637 120 + 1 GGGCGCUGCACGCAUCGAGUCGGUGGACACCACCACUGUAUAGUACGACCAAGAGCUUGGUGCAGCCAACAAAAUGUUUAGGCGGAUGUCCGCCAGCAGUAGCAUCGCGUUUGGAGUUCA (((((((((((((.((..((((((((...)))))(((....)))..)))...))))...))))))).....((((((...(((((....))))).((....))...))))))...)))). ( -43.10) >DroMoj_CAF1 1753 117 + 1 UGGCGCUUCACGUAUUGAGUCUGUGGACACAACCACAGUAUAGUAUGACCAAUUCGGUAGCGCUGCCACAAAAAGGCCACGGCGGAUGUCCGCCGCCAGC---GCCACAUGUGGAGUUCA .((((((...(((((((...((((((......))))))..)))))))(((.....))))))))).(((((....((((.((((((....))))))...).---)))...)))))...... ( -47.30) >DroAna_CAF1 1815 117 + 1 GGGUGGUGCUCGUAUCGAAUCUGUGGACACCACCACAGUAUAGUAUGACCCAGCAGUCAGCUCAGCCACAAAAUGGUUGCGGCGGAUGUCCGCCGGCAGC---AUAACGUUUGGAGUUCA ((((.(((((.(((......((((((......)))))))))))))).))))(((.....))).(((..((((...((((((((((....))))).)))))---......))))..))).. ( -43.00) >DroPer_CAF1 1808 117 + 1 GAGCGCUGCACGUAUCGAGUCUGUGGACACCACCACAGUAUAGUAUGACCAAGCGCUUAGCGCAGCCACCAAACGGCUGCGGCGGAUGUCCGCCUGCAGU---GCCACGCAUGGAGUUCA ((((.((((.(((((.....((((((......))))))....))))).....(((((..((((((((.......))))))(((((....))))).)))))---))...))).)..)))). ( -49.20) >consensus GGGCGCUGCACGUAUCGAGUCUGUGGACACCACCACAGUAUAGUAUGACCAAGCGCUUAGCGCAGCCACCAAACGGCUGCGGCGGAUGUCCGCCGGCAGC___GCCACGUUUGGAGUUCA ((((......(((((.....((((((......))))))....))))).(((.(((....((((((((.......))))))(((((....))))).........))..))).))).)))). (-30.58 = -31.42 + 0.84)

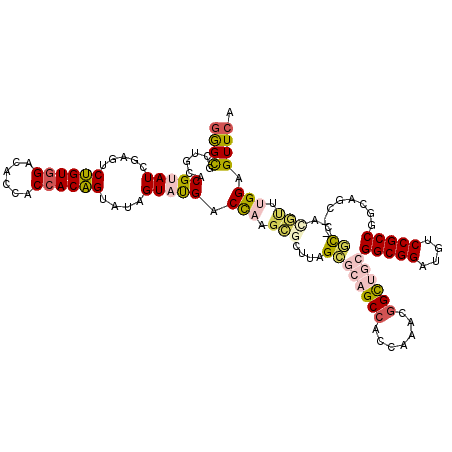
| Location | 231,974 – 232,089 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -20.39 |
| Energy contribution | -20.87 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 231974 115 - 27905053 U-CCCGAUUUUUUCUAAACAAAAUGUUACUUUUUUUCGUUGAACUCCAAACAUUGCGCUGUCGGCGGAAGUCCGCCGCGACCGUUCAUUGGCUGCGCCAAGCGCAAAGUCAUACUA .-...((((((..........((((((.....(((.....))).....))))))(((((...(((((....)))))(((.(((.....))).)))....)))))))))))...... ( -30.40) >DroVir_CAF1 1806 115 - 1 U-UUCUGUUGCGUCUAAACAAAAUGUUACUCUUUUUCGUUGAACUCCACACGUGGCGCUGGCGGCGGACAUCCGCCUUGGCCGUUUUGUGGCAGAACUACCAACAUGAUCAUACUA .-.(((((((..(((..((((((((....................(((....))).((..(.(((((....))))))..))))))))))...))).....))))).))........ ( -30.00) >DroPse_CAF1 1848 115 - 1 U-CUCGGCUUUUUCUAAACAAAAUGUUACUUUUUUUCGUUGAACUCCAUGCGUGGCACUGCAGGCGGACAUCCGCCGCAGCCGUUUGGUGGCUGCGCUAAGCGCUUGGUCAUACUA .-.(((((........(((.....)))..........)))))...(((.(((((((......(((((....)))))(((((((.....))))))))))..)))).)))........ ( -37.37) >DroMoj_CAF1 1793 115 - 1 U-UUGUGUUGUGUCUAAACAAAAUGUUACUCUUUUUCGUUGAACUCCACAUGUGGCGCUGGCGGCGGACAUCCGCCGUGGCCUUUUUGUGGCAGCGCUACCGAAUUGGUCAUACUA .-.....((((......))))...((....((..(((((.(....).))..((((((((.(((((((....))))))).(((.......))))))))))).)))..))....)).. ( -37.30) >DroAna_CAF1 1855 116 - 1 UUUUAGACUUUCUCUAAACAAAAUGUUACUCUUUUUCGAUGAACUCCAAACGUUAUGCUGCCGGCGGACAUCCGCCGCAACCAUUUUGUGGCUGAGCUGACUGCUGGGUCAUACUA ..(((((.....)))))((((((((............(...(((.......)))...)(((.(((((....))))))))..))))))))((((.(((.....))).))))...... ( -31.90) >DroPer_CAF1 1848 115 - 1 U-CUCGGCUUUUUCUAAACAAAAUGUUACUUUUUUUCGUUGAACUCCAUGCGUGGCACUGCAGGCGGACAUCCGCCGCAGCCGUUUGGUGGCUGCGCUAAGCGCUUGGUCAUACUA .-.(((((........(((.....)))..........)))))...(((.(((((((......(((((....)))))(((((((.....))))))))))..)))).)))........ ( -37.37) >consensus U_CUCGGCUUUUUCUAAACAAAAUGUUACUCUUUUUCGUUGAACUCCAAACGUGGCGCUGCCGGCGGACAUCCGCCGCAGCCGUUUUGUGGCUGCGCUAACCGCUUGGUCAUACUA ...................................................(((((......(((((....)))))(((((((.....)))))))............))))).... (-20.39 = -20.87 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:40 2006