
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,257,116 – 7,257,300 |
| Length | 184 |
| Max. P | 0.969423 |

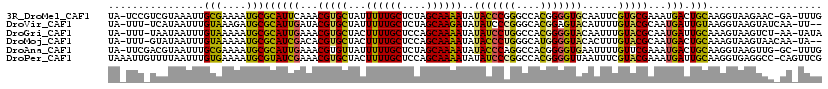
| Location | 7,257,116 – 7,257,232 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -19.24 |
| Energy contribution | -18.22 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7257116 116 - 27905053 UA-UCCGUCGUAAAUUGCGAAAAUGCGCAUUCAAACGUGCUAUUUUUGCUCUAGCAAAAUAUACCCGGGCCACGGGGUGCAAUUCGUGCGAAAUGACUGCAAGGUAAGAAC-GA-UUUG ..-...(((((.....((((((((((((........)))).))))))))(((.((........((((.....)))).((((..((((.....)))).))))..)).)))))-))-)... ( -32.60) >DroVir_CAF1 1246 114 - 1 UA-UUU-UCAUAAUUUGUAAAGAUGCGCAUUGAUACGUGCUAUUUUUGCUCUAGCAAGAUAUAUCCCGGGCACGGAGUACAUUUUGUACGCAAUGAUUGUAAGGUAAGUAUCAA-UU-- ((-(..-((((.....((((((((((((........)))).))))))))....(((((((.((..(((....)))..)).))))))).....))))..))).............-..-- ( -26.40) >DroGri_CAF1 1274 115 - 1 UA-UUU-UAAUAAUUUGUAAAAAUGCGCAUUGAAACGUGCUACUUUUGCUCCAGCAAAAUAUAUCCUGGCCACGGGGUACAAUUUGUACGCAAUGAUUGCAAAGUAAGUCU-AA-UAUA ..-...-................(((.(((((...(((((...((((((....))))))..(((((((....)))))))......))))))))))...)))..........-..-.... ( -24.80) >DroMoj_CAF1 566 114 - 1 UA-UUU-GUAUAAUUUGUAAAAAUGCGCAUCGACACGUGCUACUUUUGCUCCAGCAAAAUAUACCCUGGGCAUGGGGUACACUUUGUACGCAAUGACUGCAAAGUAAGUAACAA-UA-- ((-(((-((((((..((((.....((((........))))..((..(((.((((...........)))))))..)).))))..))))))(((.....))).)))))........-..-- ( -25.60) >DroAna_CAF1 1211 116 - 1 UA-UUCGACGUAAUUUGCGAAAAUGCGCAUUGAAACGUGUUAUUUUUGCUCUAGCAAAAUAUACCCAGGCCACGGGGUGAAUUUUGUUCGAAAUGACUGCAAGGUAAGUUG-GC-UUUG ..-(((((........((((((((((((........))).)))))))))....(((((((.(((((.(....).))))).)))))))))))).......((((((......-))-)))) ( -31.30) >DroPer_CAF1 1249 118 - 1 UAAAUUGUUUUAAUUUGUGAAAAUGCGUAUCGAAACGUGCUACUUUUGCUCCAGCAAAAUAUAUCCCGGCCACGGGGUUAAUUUCGUACGAAAUGAUUGCAAGGUGAGGCC-CAGUUCG ......((((((.((((..(.....((((.(((((........((((((....))))))...((((((....))))))...)))))))))......)..)))).)))))).-....... ( -29.40) >consensus UA_UUU_UCAUAAUUUGUAAAAAUGCGCAUUGAAACGUGCUACUUUUGCUCCAGCAAAAUAUACCCCGGCCACGGGGUACAUUUUGUACGAAAUGACUGCAAGGUAAGUAC_AA_UUUG ................(((....)))((((((...(((((...((((((....))))))..(((((((....)))))))......)))))...))).)))................... (-19.24 = -18.22 + -1.02)


| Location | 7,257,154 – 7,257,271 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -17.75 |
| Energy contribution | -18.03 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7257154 117 + 27905053 CACCCCGUGGCCCGGGUAUAUUUUGCUAGAGCAAAAAUAGCACGUUUGAAUGCGCAUUUUCGCAAUUUACGACGGA-UAAAUUACGAAAACAAAACAAAUGCGCA-AGCUGCUAGCACG ...((((.....)))).......(((((((((..((((.....))))...((((((((((((.(((((((....).-)))))).))).........)))))))))-.))).)))))).. ( -30.60) >DroVir_CAF1 1283 111 + 1 UACUCCGUGCCCGGGAUAUAUCUUGCUAGAGCAAAAAUAGCACGUAUCAAUGCGCAUCUUUACAAAUUAUGA-AAA-UAACUUGUAAAAAU-----AAA-ACGCAAUGCUGUCAGCACG .....(((((.((((((((....(((((.........))))).)))))..((((.((.(((((((.((((..-..)-))).))))))).))-----...-.))))...)))...))))) ( -26.00) >DroPse_CAF1 1297 111 + 1 AACCCCGUGGCCGGGAUAUAUUUUGCUGGAGCAAAAGUAGCACGUUUCGAUACGCAUUUUCACAAAUUAAAACAAUUUAAUUUGCGAAAA----ACAAA-GCGCA--GC-ACUAGCACG ...((((....))))........((((((.((....((.((.(((......)))..(((((.(((((((((....))))))))).)))))----.....-)))).--))-.)))))).. ( -27.90) >DroGri_CAF1 1312 111 + 1 UACCCCGUGGCCAGGAUAUAUUUUGCUGGAGCAAAAGUAGCACGUUUCAAUGCGCAUUUUUACAAAUUAUUA-AAA-UAACUUGUAAAAAU-----AAA-ACGCAAAACAGUCAGCACA ....((.......))........((((((.((.......))..((((...((((.((((((((((.((((..-..)-))).))))))))))-----...-.))))))))..)))))).. ( -22.40) >DroAna_CAF1 1249 115 + 1 CACCCCGUGGCCUGGGUAUAUUUUGCUAGAGCAAAAAUAACACGUUUCAAUGCGCAUUUUCGCAAAUUACGUCGAA-UAAUUUGCGAAAA-AUAACAAA-ACGCA-GGCAGCAAGCACG ......((.(((((.((...((((((....))))))......(((......)))..(((((((((((((.......-)))))))))))))-........-)).))-))).))....... ( -32.70) >DroPer_CAF1 1288 111 + 1 AACCCCGUGGCCGGGAUAUAUUUUGCUGGAGCAAAAGUAGCACGUUUCGAUACGCAUUUUCACAAAUUAAAACAAUUUAAUUUGCGAAAA----ACAAA-GCGCA--GC-ACUAGCACG ...((((....))))........((((((.((....((.((.(((......)))..(((((.(((((((((....))))))))).)))))----.....-)))).--))-.)))))).. ( -27.90) >consensus AACCCCGUGGCCGGGAUAUAUUUUGCUAGAGCAAAAAUAGCACGUUUCAAUGCGCAUUUUCACAAAUUAAGACAAA_UAAUUUGCGAAAA____ACAAA_ACGCA__GC_GCUAGCACG ......(((...........((((((....))))))...((.(((......)))..(((((((((((((........)))))))))))))............))...........))). (-17.75 = -18.03 + 0.28)



| Location | 7,257,154 – 7,257,271 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -29.93 |
| Energy contribution | -27.99 |
| Covariance contribution | -1.94 |
| Combinations/Pair | 1.42 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7257154 117 - 27905053 CGUGCUAGCAGCU-UGCGCAUUUGUUUUGUUUUCGUAAUUUA-UCCGUCGUAAAUUGCGAAAAUGCGCAUUCAAACGUGCUAUUUUUGCUCUAGCAAAAUAUACCCGGGCCACGGGGUG ..((((((.(((.-.((((.((((...(((((((((((((((-(.....)))))))))))))....)))..)))).)))).......))))))))).....(((((.(....).))))) ( -39.50) >DroVir_CAF1 1283 111 - 1 CGUGCUGACAGCAUUGCGU-UUU-----AUUUUUACAAGUUA-UUU-UCAUAAUUUGUAAAGAUGCGCAUUGAUACGUGCUAUUUUUGCUCUAGCAAGAUAUAUCCCGGGCACGGAGUA ((((((.....((.(((((-...-----((((((((((((((-(..-..)))))))))))))))))))).))...((.(.(((((((((....)))))).))).).))))))))..... ( -35.50) >DroPse_CAF1 1297 111 - 1 CGUGCUAGU-GC--UGCGC-UUUGU----UUUUCGCAAAUUAAAUUGUUUUAAUUUGUGAAAAUGCGUAUCGAAACGUGCUACUUUUGCUCCAGCAAAAUAUAUCCCGGCCACGGGGUU .(..(...(-(.--(((((-.....----(((((((((((((((....))))))))))))))).))))).))....)..)...((((((....))))))...((((((....)))))). ( -36.90) >DroGri_CAF1 1312 111 - 1 UGUGCUGACUGUUUUGCGU-UUU-----AUUUUUACAAGUUA-UUU-UAAUAAUUUGUAAAAAUGCGCAUUGAAACGUGCUACUUUUGCUCCAGCAAAAUAUAUCCUGGCCACGGGGUA ..(((((...(((((((((-...-----((((((((((((((-(..-..))))))))))))))))))))...))))(.((.......)).)))))).....(((((((....))))))) ( -34.40) >DroAna_CAF1 1249 115 - 1 CGUGCUUGCUGCC-UGCGU-UUUGUUAU-UUUUCGCAAAUUA-UUCGACGUAAUUUGCGAAAAUGCGCAUUGAAACGUGUUAUUUUUGCUCUAGCAAAAUAUACCCAGGCCACGGGGUG ....((((..(((-((.((-(((((...-(((((((((((((-(.....))))))))))))))...)))...))))((((...((((((....)))))).)))).)))))..))))... ( -36.30) >DroPer_CAF1 1288 111 - 1 CGUGCUAGU-GC--UGCGC-UUUGU----UUUUCGCAAAUUAAAUUGUUUUAAUUUGUGAAAAUGCGUAUCGAAACGUGCUACUUUUGCUCCAGCAAAAUAUAUCCCGGCCACGGGGUU .(..(...(-(.--(((((-.....----(((((((((((((((....))))))))))))))).))))).))....)..)...((((((....))))))...((((((....)))))). ( -36.90) >consensus CGUGCUAGC_GC__UGCGC_UUUGU____UUUUCGCAAAUUA_UUUGUCAUAAUUUGUGAAAAUGCGCAUUGAAACGUGCUACUUUUGCUCCAGCAAAAUAUAUCCCGGCCACGGGGUA .(..(.........(((((..........(((((((((((((........))))))))))))).))))).......)..)...((((((....))))))..(((((((....))))))) (-29.93 = -27.99 + -1.94)



| Location | 7,257,194 – 7,257,300 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.02 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -17.54 |
| Energy contribution | -17.65 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7257194 106 + 27905053 CACGUUUGAAUGCGCAUUUUCGCAAUUUACGACGGA-UAAAUUACGAAAACAAAACAAAUGCGCAAGCUGCUAGCACGUGUGCUGCUGAAGAAGAAGAAAUAUGAAA ((..(((...((((((((((((.(((((((....).-)))))).))).........)))))))))....((.((((....))))))...........)))..))... ( -21.60) >DroPse_CAF1 1337 93 + 1 CACGUUUCGAUACGCAUUUUCACAAAUUAAAACAAUUUAAUUUGCGAAAA----ACAAA-GCGCA-GC-ACUAGCACGUGUGUGGCUGAAGAAGAAGAGA------- ....((((....(((.(((((.(((((((((....))))))))).)))))----.....-)))((-((-....(((....))).))))..))))......------- ( -20.50) >DroEre_CAF1 1320 104 + 1 CACGUUUCAAUGCGCAUUUUCGCAAAUUACGACGAA-UAAUUUGCGAAA--AAAACAAAGGCGCAAACAGCUAGCACGUGUGCAGCUGAAGAAGAAGAAAGUUGAAA ....(((((((((((.(((((((((((((.......-))))))))))))--)........))))...(((((.(((....))))))))............))))))) ( -33.40) >DroYak_CAF1 1292 105 + 1 CACGUUUCAAUGCGCAUUUUCGCAAAUUACGACGAA-UAAUUUGCGAAAA-AAAACAAAAGCGCAAGCAGCUAGCACGUGUGCUGCUGAAGAAGAAGAGAGAUGAAA ..((((((..(((((.(((((((((((((.......-)))))))))))))-.........)))))..((((.((((....))))))))..........))))))... ( -34.70) >DroAna_CAF1 1289 104 + 1 CACGUUUCAAUGCGCAUUUUCGCAAAUUACGUCGAA-UAAUUUGCGAAAA-AUAACAAA-ACGCAGGCAGCAAGCACGUGUGCUGCUGAAGAAGAAGAACGAGGAAA ..(((((...((((..(((((((((((((.......-)))))))))))))-........-.))))..(((((.(((....))))))))........)))))...... ( -28.90) >DroPer_CAF1 1328 93 + 1 CACGUUUCGAUACGCAUUUUCACAAAUUAAAACAAUUUAAUUUGCGAAAA----ACAAA-GCGCA-GC-ACUAGCACGUGUGCGGCUGAAGAAGAAGAGA------- ....((((....(((.(((((.(((((((((....))))))))).)))))----.....-)))((-((-....(((....))).))))..))))......------- ( -22.10) >consensus CACGUUUCAAUGCGCAUUUUCGCAAAUUACGACGAA_UAAUUUGCGAAAA_AAAACAAA_GCGCAAGCAGCUAGCACGUGUGCUGCUGAAGAAGAAGAAAGAUGAAA ....(((((..((((((((((((((((((........)))))))))))).............((.........))..))))))...)))))................ (-17.54 = -17.65 + 0.11)


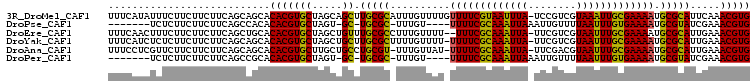
| Location | 7,257,194 – 7,257,300 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.02 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -22.43 |
| Energy contribution | -21.47 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.82 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7257194 106 - 27905053 UUUCAUAUUUCUUCUUCUUCAGCAGCACACGUGCUAGCAGCUUGCGCAUUUGUUUUGUUUUCGUAAUUUA-UCCGUCGUAAAUUGCGAAAAUGCGCAUUCAAACGUG .....................((((((....)))).)).(..(((((((((.........((((((((((-(.....))))))))))))))))))))..)....... ( -28.00) >DroPse_CAF1 1337 93 - 1 -------UCUCUUCUUCUUCAGCCACACACGUGCUAGU-GC-UGCGC-UUUGU----UUUUCGCAAAUUAAAUUGUUUUAAUUUGUGAAAAUGCGUAUCGAAACGUG -------....................(((((.....(-(.-(((((-.....----(((((((((((((((....))))))))))))))).))))).))..))))) ( -26.30) >DroEre_CAF1 1320 104 - 1 UUUCAACUUUCUUCUUCUUCAGCUGCACACGUGCUAGCUGUUUGCGCCUUUGUUUU--UUUCGCAAAUUA-UUCGUCGUAAUUUGCGAAAAUGCGCAUUGAAACGUG ((((((.............((((((((....))).)))))..(((((........(--((((((((((((-(.....)))))))))))))).))))))))))).... ( -33.20) >DroYak_CAF1 1292 105 - 1 UUUCAUCUCUCUUCUUCUUCAGCAGCACACGUGCUAGCUGCUUGCGCUUUUGUUUU-UUUUCGCAAAUUA-UUCGUCGUAAUUUGCGAAAAUGCGCAUUGAAACGUG (((((..............((((((((....)))).))))..(((((.........-(((((((((((((-(.....)))))))))))))).))))).))))).... ( -32.70) >DroAna_CAF1 1289 104 - 1 UUUCCUCGUUCUUCUUCUUCAGCAGCACACGUGCUUGCUGCCUGCGU-UUUGUUAU-UUUUCGCAAAUUA-UUCGACGUAAUUUGCGAAAAUGCGCAUUGAAACGUG ...................((((((((....))).)))))...((((-(((((...-(((((((((((((-(.....))))))))))))))...)))...)))))). ( -30.90) >DroPer_CAF1 1328 93 - 1 -------UCUCUUCUUCUUCAGCCGCACACGUGCUAGU-GC-UGCGC-UUUGU----UUUUCGCAAAUUAAAUUGUUUUAAUUUGUGAAAAUGCGUAUCGAAACGUG -------....................(((((.....(-(.-(((((-.....----(((((((((((((((....))))))))))))))).))))).))..))))) ( -26.30) >consensus UUUCAUCUCUCUUCUUCUUCAGCAGCACACGUGCUAGCUGCUUGCGC_UUUGUUUU_UUUUCGCAAAUUA_UUCGUCGUAAUUUGCGAAAAUGCGCAUUGAAACGUG ...........................(((((((.....)).(((((..........(((((((((((((........))))))))))))).))))).....))))) (-22.43 = -21.47 + -0.97)
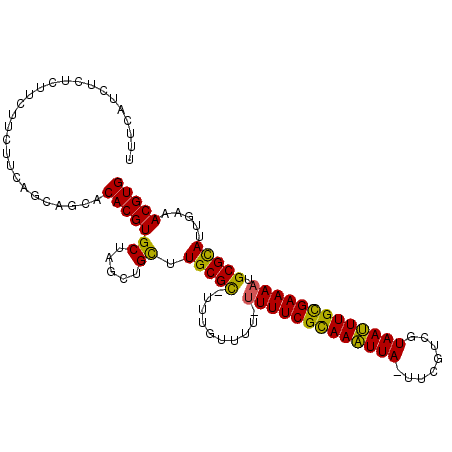

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:32 2006