
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,236,542 – 7,236,689 |
| Length | 147 |
| Max. P | 0.980941 |

| Location | 7,236,542 – 7,236,650 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.92 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -23.31 |
| Energy contribution | -23.28 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7236542 108 + 27905053 -UAAGUCUGCUAUUCAUUUCUUAUGGG-UUUUAAUGUGCGAAGUCUGUUCCCCUGUUUGGCGUUCAAUUUUCGCCUGA-AAGGUUUUAAGACUUCGUAGAUCAAAUAUAAG -...((((((.((((((.....)))))-)..........(((((((.....(((....((((.........))))...-.))).....))))))))))))).......... ( -28.20) >DroSec_CAF1 686 109 + 1 -UCUGUCUAUUAUUCAUUUCUUAGGGU-UUUCAAUGUGCGAAGUCUGUUCCCCUGUUUGGCGUUCAAUUUUCGCCUAAAAAGGUUUUAAGACUUCGCAGAUGAAAUAUGAG -..................((((....-(((((.(.((((((((((.....(((.(((((((.........)))).))).))).....)))))))))).))))))..)))) ( -31.80) >DroSim_CAF1 693 108 + 1 -UCUGUCUGUUAUUCAUUUCGUAGGGU-UUUCAAUGUGCGAAGUCUGUUCCCCUGUUUGGCGUUCAAUUUUCGCCUGA-AAGGUUUUAAGACUUCGCAGAUCAAAUAUGAG -.................(((((.(((-(.......((((((((((.....(((....((((.........))))...-.))).....))))))))))))))...))))). ( -30.71) >DroEre_CAF1 691 108 + 1 UACAAUUUUUCAGUAAUUUCUUAAGGU-UUUCAAUAUGCGAAGUC-GUUCCCCUCAUUGGCGUUCAAUUUUCGCCUGU-AAGGUUUUAAGACUCCGCAGAACAAUUAUAAG ...................((((..((-((((....((((.((((-.....(((.((.((((.........)))).))-.)))......)))).))))))).)))..)))) ( -21.70) >DroYak_CAF1 698 109 + 1 UUUAAUUUUUGAGUAAUUUCUUAGGUU-UUUUAAUGUGCGAAAUCUGUUCCCCUCGUUGGCGUUCAAUUUUCGCCUGU-AAGGUUUUAAGAUUUCGCAGAUCAAAUAUAAG .......((((((......))))))..-.(((.((.((((((((((.....(((..(.((((.........)))).).-.))).....)))))))))).)).)))...... ( -28.00) >DroPer_CAF1 1041 99 + 1 UAUAGU-----AGA----ACGUAUGUAUAUUU--UAUGCGAAGUCUGUUCCCCUCCGUGGCGUCCAGAGAUCGCCUUG-GAGGUUUAAGUACUUCGCAGAGCAACUGUAGC ((((((-----...----((....))......--..((((((((((.....((((((.((((((....)).)))).))-))))....)).)))))))).....)))))).. ( -33.40) >consensus _UCAGUCUGUUAGUCAUUUCUUAGGGU_UUUCAAUGUGCGAAGUCUGUUCCCCUCUUUGGCGUUCAAUUUUCGCCUGA_AAGGUUUUAAGACUUCGCAGAUCAAAUAUAAG ....................................((((((((((.....(((....((((.........)))).....))).....))))))))))............. (-23.31 = -23.28 + -0.02)


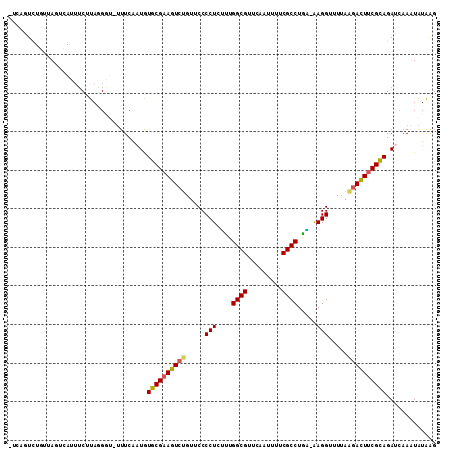
| Location | 7,236,542 – 7,236,650 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.92 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -22.78 |
| Energy contribution | -23.62 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7236542 108 - 27905053 CUUAUAUUUGAUCUACGAAGUCUUAAAACCUU-UCAGGCGAAAAUUGAACGCCAAACAGGGGAACAGACUUCGCACAUUAAAA-CCCAUAAGAAAUGAAUAGCAGACUUA- (((((.((((((...((((((((.....(((.-...((((.........))))....))).....))))))))...)))))).-...)))))..................- ( -22.90) >DroSec_CAF1 686 109 - 1 CUCAUAUUUCAUCUGCGAAGUCUUAAAACCUUUUUAGGCGAAAAUUGAACGCCAAACAGGGGAACAGACUUCGCACAUUGAAA-ACCCUAAGAAAUGAAUAAUAGACAGA- ......(((((..((((((((((.....(((.(((.((((.........))))))).))).....))))))))))...)))))-..........................- ( -29.40) >DroSim_CAF1 693 108 - 1 CUCAUAUUUGAUCUGCGAAGUCUUAAAACCUU-UCAGGCGAAAAUUGAACGCCAAACAGGGGAACAGACUUCGCACAUUGAAA-ACCCUACGAAAUGAAUAACAGACAGA- ......((..((.((((((((((.....(((.-...((((.........))))....))).....)))))))))).))..)).-..........................- ( -27.90) >DroEre_CAF1 691 108 - 1 CUUAUAAUUGUUCUGCGGAGUCUUAAAACCUU-ACAGGCGAAAAUUGAACGCCAAUGAGGGGAAC-GACUUCGCAUAUUGAAA-ACCUUAAGAAAUUACUGAAAAAUUGUA ..(((((((.((((((((((((......((((-(..((((.........))))..))))).....-)))))))))........-....(((....)))..))).))))))) ( -29.70) >DroYak_CAF1 698 109 - 1 CUUAUAUUUGAUCUGCGAAAUCUUAAAACCUU-ACAGGCGAAAAUUGAACGCCAACGAGGGGAACAGAUUUCGCACAUUAAAA-AACCUAAGAAAUUACUCAAAAAUUAAA ......((((((.((((((((((.....((((-...((((.........))))...)))).....)))))))))).)))))).-........................... ( -27.50) >DroPer_CAF1 1041 99 - 1 GCUACAGUUGCUCUGCGAAGUACUUAAACCUC-CAAGGCGAUCUCUGGACGCCACGGAGGGGAACAGACUUCGCAUA--AAAUAUACAUACGU----UCU-----ACUAUA ((.......))..((((((((.......((((-(..((((.((....))))))..))))).......))))))))..--..............----...-----...... ( -30.64) >consensus CUUAUAUUUGAUCUGCGAAGUCUUAAAACCUU_UCAGGCGAAAAUUGAACGCCAAACAGGGGAACAGACUUCGCACAUUAAAA_ACCCUAAGAAAUGAAUAAAAGACUGA_ .......(((((.((((((((((.....((((....((((.........))))....))))....)))))))))).))))).............................. (-22.78 = -23.62 + 0.84)



| Location | 7,236,574 – 7,236,689 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.54 |
| Mean single sequence MFE | -32.14 |
| Consensus MFE | -25.15 |
| Energy contribution | -24.65 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7236574 115 + 27905053 UGUGCGAAGUCUGUUCCCCUGUUUGGCGUUCAAUUUUCGCCUGA-AAGGUUUUAAGACUUCGUAGAUCAAAUAUA---AGCAA-UCAUUCACCCGCAUCCAAGUGAAGCUCAAUUGGUUC ..((((((((((.....(((....((((.........))))...-.))).....))))))))))((((((.....---(((..-...(((((..........))))))))...)))))). ( -30.30) >DroSec_CAF1 718 116 + 1 UGUGCGAAGUCUGUUCCCCUGUUUGGCGUUCAAUUUUCGCCUAAAAAGGUUUUAAGACUUCGCAGAUGAAAUAUG---AGCAA-UCAUUCACCCGCAUCCAAGUGAAGCUCAAUUGGUUC ..((((((((((.....(((.(((((((.........)))).))).))).....)))))))))).........((---(((..-...(((((..........))))))))))........ ( -34.60) >DroEre_CAF1 724 114 + 1 UAUGCGAAGUC-GUUCCCCUCAUUGGCGUUCAAUUUUCGCCUGU-AAGGUUUUAAGACUCCGCAGAACAAUUAUA---AGCAA-UCAUUCACCCGCGUCCAAGUGAAGCUCAAUUGGUUC ..((((.((((-.....(((.((.((((.........)))).))-.)))......)))).))))((((.......---(((..-...(((((..........))))))))......)))) ( -26.22) >DroWil_CAF1 1264 118 + 1 UGUGCGAAACCUGUUCCCCUUGCUGGCGUGCAGUGAUAGCCUUC-AAGGUUUUAAGGUUUCGCAAAUCAAAUGUACAGAGCAA-AAAUUCACCCGCCUCCAAGUGAAGCACUAUUGGUCA ..((((((((((.....(((((..(((...........)))..)-)))).....)))))))))).............((.(((-...(((((..........)))))......))).)). ( -34.70) >DroYak_CAF1 731 115 + 1 UGUGCGAAAUCUGUUCCCCUCGUUGGCGUUCAAUUUUCGCCUGU-AAGGUUUUAAGAUUUCGCAGAUCAAAUAUA---AGCAA-UCAUUCACCCGCGUCCAAGUGAAGCACAAUUGGUUC ..((((((((((.....(((..(.((((.........)))).).-.))).....))))))))))..........(---(.(((-(..(((((..........))))).....)))).)). ( -28.60) >DroMoj_CAF1 74633 114 + 1 -AUGCGACAUUUGUUCCCCUUGCUGGCGUGCAGCUGUAGCCAGA-AAGGUUUCAAGAUGUCGCAGAGCAAACA-G---AGCAACUGUUUCACCCGCAUCCAACUGAAGUACUAUUGGGUU -.((((((((((.....((((.(((((.(((....)))))))).-)))).....))))))))))..(((((((-(---.....)))))).....))((((((..(.....)..)))))). ( -38.40) >consensus UGUGCGAAAUCUGUUCCCCUCGUUGGCGUUCAAUUUUCGCCUGA_AAGGUUUUAAGACUUCGCAGAUCAAAUAUA___AGCAA_UCAUUCACCCGCAUCCAAGUGAAGCACAAUUGGUUC ..((((((((((.....(((....((((.........)))).....))).....))))))))))................(((....(((((..........)))))......))).... (-25.15 = -24.65 + -0.50)



| Location | 7,236,574 – 7,236,689 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.54 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -28.44 |
| Energy contribution | -28.25 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7236574 115 - 27905053 GAACCAAUUGAGCUUCACUUGGAUGCGGGUGAAUGA-UUGCU---UAUAUUUGAUCUACGAAGUCUUAAAACCUU-UCAGGCGAAAAUUGAACGCCAAACAGGGGAACAGACUUCGCACA ....(((.(((((((((((((....))))))))...-..)))---))...))).....((((((((.....(((.-...((((.........))))....))).....)))))))).... ( -31.90) >DroSec_CAF1 718 116 - 1 GAACCAAUUGAGCUUCACUUGGAUGCGGGUGAAUGA-UUGCU---CAUAUUUCAUCUGCGAAGUCUUAAAACCUUUUUAGGCGAAAAUUGAACGCCAAACAGGGGAACAGACUUCGCACA (((.....(((((((((((((....))))))))...-..)))---))...)))...((((((((((.....(((.(((.((((.........))))))).))).....)))))))))).. ( -39.50) >DroEre_CAF1 724 114 - 1 GAACCAAUUGAGCUUCACUUGGACGCGGGUGAAUGA-UUGCU---UAUAAUUGUUCUGCGGAGUCUUAAAACCUU-ACAGGCGAAAAUUGAACGCCAAUGAGGGGAAC-GACUUCGCAUA ((((....(((((((((((((....))))))))...-..)))---)).....))))(((((((((......((((-(..((((.........))))..))))).....-))))))))).. ( -39.80) >DroWil_CAF1 1264 118 - 1 UGACCAAUAGUGCUUCACUUGGAGGCGGGUGAAUUU-UUGCUCUGUACAUUUGAUUUGCGAAACCUUAAAACCUU-GAAGGCUAUCACUGCACGCCAGCAAGGGGAACAGGUUUCGCACA ......((((.((((((((((....))))))))...-..)).))))..........((((((((((.....((((-(..(((...........)))..))))).....)))))))))).. ( -38.90) >DroYak_CAF1 731 115 - 1 GAACCAAUUGUGCUUCACUUGGACGCGGGUGAAUGA-UUGCU---UAUAUUUGAUCUGCGAAAUCUUAAAACCUU-ACAGGCGAAAAUUGAACGCCAACGAGGGGAACAGAUUUCGCACA ....((((..(...(((((((....))))))))..)-)))..---...........((((((((((.....((((-...((((.........))))...)))).....)))))))))).. ( -33.10) >DroMoj_CAF1 74633 114 - 1 AACCCAAUAGUACUUCAGUUGGAUGCGGGUGAAACAGUUGCU---C-UGUUUGCUCUGCGACAUCUUGAAACCUU-UCUGGCUACAGCUGCACGCCAGCAAGGGGAACAAAUGUCGCAU- ...(((((.(.....).)))))....((((.((((((.....---)-)))))))))((((((((.(((...((((-((((((...........))))).)))))...))))))))))).- ( -39.10) >consensus GAACCAAUUGAGCUUCACUUGGAUGCGGGUGAAUGA_UUGCU___UAUAUUUGAUCUGCGAAAUCUUAAAACCUU_UCAGGCGAAAAUUGAACGCCAACAAGGGGAACAGACUUCGCACA .............((((((((....)))))))).......................((((((((((.....((((....((((.........))))....))))....)))))))))).. (-28.44 = -28.25 + -0.19)

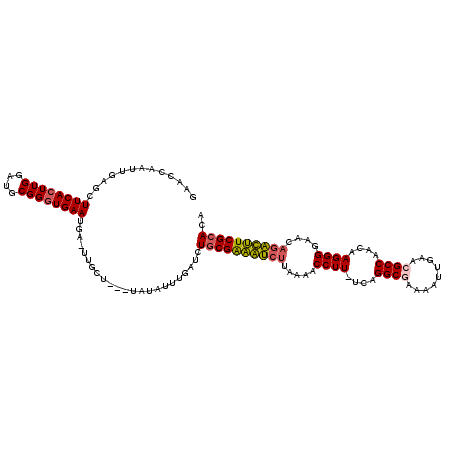
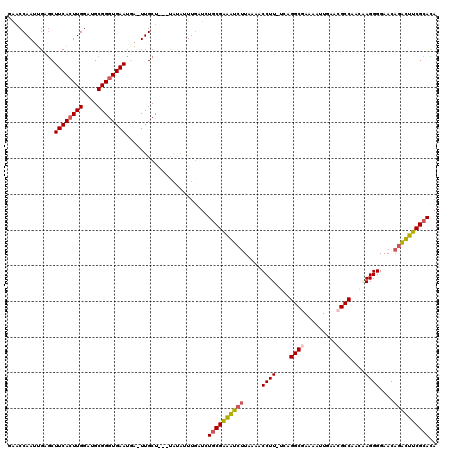
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:22 2006